Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4706
Review Article(ISSN: 2637-4706) 
Intrinsically Disordered Proteins (IDPs): Experimental and Computational Approaches in Drug Discovery Volume 1 - Issue 5
Suresh Kumar*
- Department of Diagnostic and Allied Health Science, Faculty of Health and Life Sciences, Management & Science University, Malaysia
Received: June 24, 2018; Published: July 02, 2018
Corresponding author:Suresh Kumar, Department of Diagnostic and Allied Health Science, Faculty of Health and Life Sciences, Management & Science University, Shah Alam, Selangor, Darul Ehsan, Malaysia
DOI: 10.32474/DDIPIJ.2018.01.000123
Abstract
Intrinsically disordered proteins (IDPs) are proteins that usually do not adopt well-defined native structures when isolated in solution under physiological conditions. Numerous IDPs have close relationships with human diseases such as Parkinson disease, Alzheimer disease, diabetes, and so on. Recently IDPs are getting importance as desirable drug targets for major diseases. This review introduces IDP’s in drug discovery by experimental and by computational approach.
Keywords: Protein disorder; Intrinsically Disordered Proteins; Disprot
Abbrevations: IDPs: Intrinsically Disordered Proteins, CD: Circular Dichroism, SAXS: Small-Angle Scattering of X-rays, PTMs: Post Translational Modifications, IDRs: Intrinsically Disordered Regions, IDPs: Intrinsically Disordered Proteins, pE-DB: Protein Ensemble Database
Introduction
Soluble proteins may consist of globular units and nonglobular units. Globular units are composed of regular secondary structure elements whereas the non-globular units are composed of disordered, unstructured, and flexible regions without regular secondary structure elements [1]. Recently it is well known that many functionally important protein segments occur outside of globular units [2,3]. Intrinsically Disordered Proteins (IDPs) (also known as intrinsically unstructured, unfolded, natively disordered or rheomorphic proteins) lack a well-defined 3D structure and exhibit a multitude of conformations that dynamically change over time and population [4]. Many proteins cannot be over-expressed, purified or crystallized. Native conformational disorder of proteins is one of the main obstacles facing structural biology analyses. Moreover, in structural genomics initiatives, it is becoming increasingly important to identify Intrinsically Disordered Proteins during the target selection process [5,6]. No commonly agreed definition of Intrinsically Disordered Proteins exists. AIDPs can be either completely unfolded or comprise both folded and unfolded segments” [7]. While completely unfolded proteins carry out their function by means of regions that lack specific 3D structure and exists as an ensemble of flexible molecules, in partially unfolded proteins only localized regions lack organized structure.
Experimental Determination of Intrinsically Disordered Proteins
IDPs are indirectly observed with a variety of experimental methods, such as X-ray crystallography, NMR, Raman spectroscopy, CD spectroscopy, and hydrodynamics measurements.
X-Ray Crystallography: Disorder leads to missing electron density in protein structures determined by X-ray crystallography. Two types of disorder have been recognized: static and dynamic [8]. Disorder is static when different molecules are rigid and adopt different conformations. It is, on the contrary, dynamic when molecules are flexible and oscillate between various conformations. If a region exists as an ensemble of φ and ψ angles, whether static or dynamic, it is intrinsically disordered. The major disadvantage of X-ray crystallography is that it requires additional experimental confirmation whether the missing electron density is a wobbly domain, is intrinsically disordered, or is the result of technical difficulties [9].
Nuclear Magnetic Resonance Spectroscopy (NMR): Protein three-dimensional structures can be determined in solution by NMR. Under favourable circumstances, NMR provides motional information on a residue-by-residue basis by means of a variety of different isotopic labelling and pulse sequence experiments [10].
Circular Dichroism (CD) Spectroscopy: Structural information for proteins in solution is also provided by circular dichroism. The circular dichroism spectroscopy can give semiquantitative information by combined use of near and far UV CD. It does not provide clear information for the proteins that contain both ordered and disordered regions [11].
Small-Angle Scattering of X-rays (SAXS): It is one of powerful method for characterization of both ordered and disordered proteins in solution. It allows one to quantitatively characterize conformational poly dispersity, in particular, of completely or partially disordered macromolecules, including multi-domain proteins with flexible linkers and intrinsically disordered proteins (IDPs) [12].
Protease Digestion
Protease digestion is a well-recognized method, which gives insight into protein structure and flexibility. The protein digestion method is particularly useful when used in combination with other methods. Protein digestion along with X-diffraction method helps to sort out whether a region of missing electron density is due to a wobbly domain or to intrinsic disorder. Protein digestion is useful when coupled with CD spectra, which lack position-specific information. Finally, the combination of proteolysis and mass spectrometry for fragment identification can indicate the presence of intrinsically disordered regions [13].
Stokes Radius Determination
Random coil disorder has also been detected by various methods for obtaining stokes radius such as small-angle X-ray scattering or size exclusion chromatography [14].
Biological Importance of Intrinsically Disordered Proteins
The native unstructured proteins are found to participate in many biological processes and commonly occur in cell signalling pathways, DNA transcription and replication and protein translation [15]. Intrinsically Disordered Proteins is important for understanding protein function as well as protein folding pathways [16]. More than 180 such proteins are known including Tau, Prions, Bcl-2, p53, 4E-BP1, elF1A and HMG proteins [17]. They are thought to become ordered when they are bound to another molecule (e.g. CREB-CBP complex) although little is understood about the cellular and structural meaning of disorder. Due to the abnormal aggregation patterns of these proteins, they are involved in major protein diseases such as Parkinson’s and Alzheimer’s syndromes [18].
Intrinsically Disordered Proteins as Drug Targets
The recent studies suggest that IDPs involved in various disease that suggests that they can be used as viable targets. IDPs including the development of small molecules that either:
(i) directly bind to the disordered ensemble, or
(ii) That bind to binding partner and inhibit IDP binding or stabilize its bound state, and
(iii) That affects modification pathways [19]. They are aimed to targeting conformational transitions and regulatory elements besides of modulating Post Translational Modifications (PTMs). Currently existing drug design strategies for IDPs include the development of small molecule and peptide inhibitors, arresting order-to-disorder transitions, targeting regulatory elements and the modulation of post-translational modifications [20].
Computational Tools for IDP’s Drug Discovery Study
Database Resource of IDP’s
A. Disprot: It is a manually curated database that provides information about proteins that lack fixed 3D structure in their putatively native states, either entirely or in part. The database contains experimentally characterized IDPs and includes functional information for many of the IDPs and regions [21]. In its first public release of February 2004, DisProt contained 154 proteins (190 disordered regions); whereas in June 2018 the database contained 803 proteins (2167 disordered regions). The database can be accessed at http://www.disport.org.
B. Database of Disordered Protein Prediction (D2P2): This database contains all the protein sequences from 1765 complete proteomes in which battery of disorder predictors and their variants, VL-XT, VSL2b, PrDOS, PV2, Espritz and IUPred, were run on [22]. The database can be accessed at http://d2p2.pro/.
C. MobiDB: MobiDB (http://mobidb.bio.unipd.it/) is a database of intrinsically disordered and mobile proteins. It contains different flavours of disorder in protein structures covering all UniProt sequences. The database features three levels of annotation: manually curated, indirect and predicted. Manually curated data is extracted from the DisProt database. Indirect data is inferred from PDB structures that are considered an indication of intrinsic disorder. Predicted data contains disorder annotations predicted for protein that lack by various predictors like ESpritz, IUPred, DisEMBL, GlobPlot, VSL2b and JRONN [23].
D. IDEAL: IDEAL (http://www.ideal.force.cs.is.nagoya-u. ac.jp/IDEAL/) provides a collection of knowledge on experimentally verified intrinsically disordered proteins (IDPs) or intrinsically disordered regions (IDRs). IDEAL contains manually curated annotations on IDPs in locations, structures, and functional sites such as protein binding regions and posttranslational modification sites together with references and structural domain assignments [24].
E. Protein Ensemble Database (pE-DB): PED3 (http:// pedb.vib.be/)is an openly accessible database for the deposition of structural information on IDP- and denatured protein ensembles based on Nuclear Magnetic Resonance (NMR) and Small-angle X-ray Scattering (SAXS) data [25].
Computational Tools for IDP’s Prediction
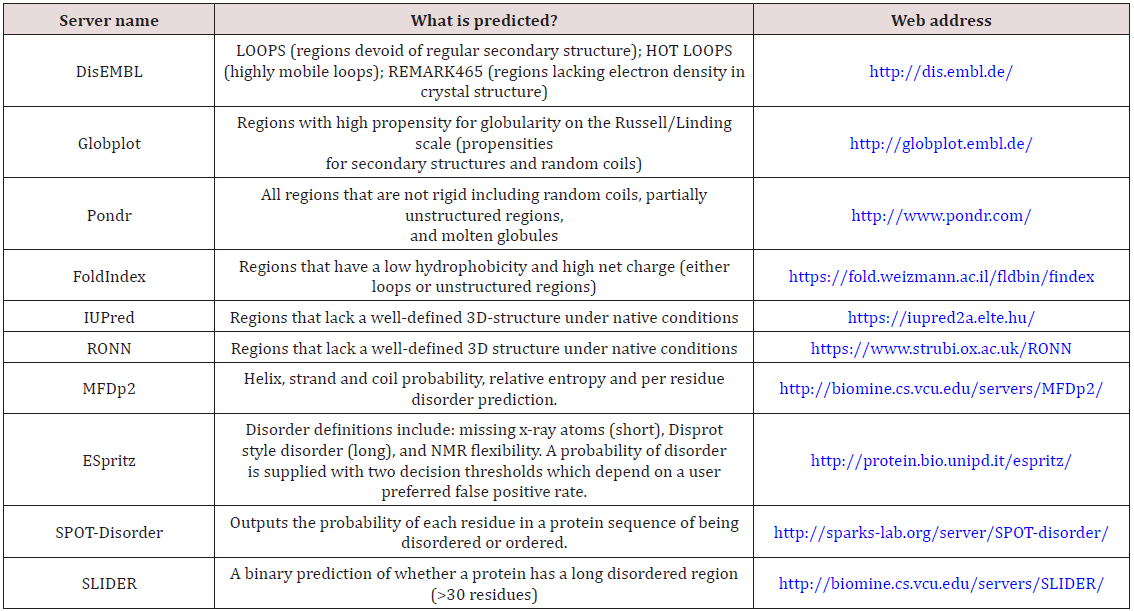
Various computational predictors were developed for predicting protein conformational disorder. Since there is no unique definition of Intrinsically Disordered Proteins, each of the predictors has its own definition and algorithms. Some of them have different versions of the same basic algorithm. Some of the predictors are based on datasets of ordered/disordered proteins. Others are based on physicochemical trends and observations. All the predictors mentioned here are available as web servers (Table 1).
Table 1: Summary of the web servers available for the prediction of protein conformational disorder.
Computational Drug Design Tools for IDPs
Experimental procedures provide a lot of useful data and computational approaches need it to carry out simulations as more realistic as possible. However, for IDPs, where dynamics is so important, computation clearly has advantages over experiments as it can describe dynamics completely and it can characterize the full conformational energy landscape of proteins. Some of docking tools for IDPs are pepATTRACT [26] and Rosetta-FlexPepDock [27] for Computational peptide protein docking. IDP-LZerD [28] used for docking of disordered protein interactions. AnchorDock [29] for Blind anchor-driven peptide docking, HADDOCK [30] used for Docking of protein-protein and protein-peptide binding. Molecular dynamics CHARMM [31] for Molecular simulations and modelling program, Amber Biomolecular simulation program, Gromacs [32] for Molecular simulations and modelling program of proteins, lipids and nucleic acids.
References
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, et al. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11(11): 1453-1459.
- Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: reassessing the protein structure-function paradigm. J MolBiol 293(2): 321-331.
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, et al. (2001) Intrinsically disordered protein. J Mol Graph Model 19(1): 26-59.
- Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, et al. (1998) Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput 473-484.
- Smialowski P, Schmidt T, Cox J, Kirschner A, Frishman D (2006) Will my protein crystallize? A sequence-based predictor. Proteins 62(2): 343- 355.
- Ferron F, Longhi S, Canard B, Karlin D (2006) A practical overview of protein disorder prediction methods. Proteins 65(1): 1-14.
- Fink AL (2005) Natively unfolded proteins. Curr Opin Struct Biol 15(1): 35-41.
- Huber R (1979) Conformational flexibility in protein molecules. Nature 280: 538-539.
- DeForte S, Uversky VN (2016) Resolving the ambiguity: making sense of intrinsic disorder when PDB structures disagree. Protein Science 25(3): 676-688.
- Tamiola K, Mulder FA (2012) Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. 40(5): 1014- 1020.
- Chemes LB, Alonso LG, Noval MG, de Prat-Gay G (2012) Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods Mol Biol 895: 387-404.
- Kikhney AG, Svergun DI (2015) A practical guide to small angle X‐ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS letters 589(19PartA): 2570-2577.
- Johnson DE, Xue B, Sickmeier MD, Meng J, Cortese MS, et al. (2012) Highthroughput characterization of intrinsic disorder in proteins from the Protein Structure Initiative. Journal of structural biology 180(1): 201- 215.
- Uversky VN (2012) Size-exclusion chromatography in structural analysis of intrinsically disordered proteins. Methods Mol Biol 896: 179-94.
- Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, et al. (2007) Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. Journal of proteome research 6(5): 1899-1916.
- Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nature reviews Molecular cell biology 16(1).
- Vilasi S, Ragone R (2008) Abundance of intrinsic disorder in SV‐IV, a multifunctional androgen‐dependent protein secreted from rat seminal vesicle. The FEBS journal 275(4): 763-774.
- Fisher CK, Stultz CM (2011) Constructing ensembles for intrinsically disordered proteins. Current opinion in structural biology 21(3): 426- 431.
- Tompa P (2012) Intrinsically disordered proteins: a 10-year recap. Trends in biochemical sciences 37(12): 509-516.
- Chen CYC, Tou WI (2013) How to design a drug for the disordered proteins?. Drug discovery today 18(19-20): 910-915.
- Piovesan D, Tabaro F, Mičetić I, Necci M, Quaglia F, et al. (2016) DisProt 7.0: a major update of the database of disordered proteins. Nucleic acids research 45(DI): D219-D227.
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, et al. (2012) D2P2: database of disordered protein predictions. Nucleic acids research 41(DI): D508-D516.
- Potenza E, Domenico TD, Walsh I, Tosatto SC (2014) MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic acids research 43(DI): D315-D320.
- Fukuchi S, Amemiya T, Sakamoto S, Nobe Y, Hosoda K, et al. (2013) IDEAL in 2014 illustrates interaction networks composed of intrinsically disordered proteins and their binding partners. Nucleic acids research 42(DI): D320-D325.
- Varadi M, Kosol S, Lebrun P, Valentini E, Blackledge M, et al. (2014) pEDB: a database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic acids research 42(DI): D326-D335.
- Schindler CE, de Vries SJ, Zacharias M (2015) Fully blind peptide-protein docking with pepATTRACT. Structure 23(8): 1507-1515.
- London N, Raveh B, Cohen E, Fathi G, Schueler Furman O (2011) Rosetta FlexPepDock web server-high resolution modeling of peptide-protein interactions. Nucleic acids research 39: W249-W253.
- Peterson LX, Roy A, Christoffer C, Terashi G, Kihara D (2017) Modeling disordered protein interactions from biophysical principles. PLoS computational biology 13(4): e1005485.
- Ben-Shimon A, Niv MY (2015) AnchorDock: blind and flexible anchordriven peptide docking. Structure 23(5): 929-940.
- DeVries SJ, Van Dijk M, Bonvin AM (2010) The HADDOCK web server for data-driven biomolecular docking. Nature protocols 5(5): 883-897.
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan SA, et al. (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. Journal of computational chemistry 4(2): 187-217.
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, et al. (2005) GROMACS: fast, flexible, and free. Journal of computational chemistry 26(16): 1701-1718.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...