Lupine Publishers Group
Lupine Publishers
Menu
Research Article(ISSN: 2641-6875) 
Preparation of Generated DNA (Streptomyces Griseus) Crown Cells (Artificial Cells) and Antibiotic Production in Its’ Co-Cultures with Yeast (Beer) Volume 1 - Issue 2
Shoshi Inooka*
- Japan Association of Science Specialists, Japan
Received: January 07, 2019; Published: January 16, 2019
*Corresponding author: Shoshi Inooka, Japan Association of Science Specialists, Japan
DOI: 10.32474/CTBM.2019.01.000108
Abstract
DNA crown cells are artificial cells which the outside of the membrane was covered of DNA. Such DNA crown cells (artificial cells) could be prepared by incubating within egg white with a sphingosine (Sph)-DNA-adenosine mixture. Also, the DNA crown cells could easy synthesized. Update, many kinds of DNA crown cells have been synthesized using various DNA. On the other hand, it is unclear whether such DNA Crown Cells. could be contribute to applied fields, Present report described that DNA crown cells using DNA of antibiotic produced Streptomyces griseus were synthesized and that the antibiotic was produced in the co-culture with yeast (beer) with egg white which contained DNA crown cells. Now, though it is studying on whether DNA crown cells were associated with the production of antibiotic, however, it was suggested that the system of the present co-cultures may be contribute to applied fields, the production of antibiotic or functional beer.
Keywords: Antibiotic; DNA Crown Cells; Sphingosine-DNA Streptomyces; Gniseus
Introduction
There has been significant progress in the generation of artificial cells since the first studies in the 1960s [1-4], yet to date, artificial cells that can replicate autonomously have not been reported. Recently, the approaches for generating fully operational (self-replicating) artificial cells have been reported [5-7]. On the mechanism on underlying the formation of artificial cells, it was demonstrated that the artificial cells were covered with DNA, (named DNA crown cells,) [8] and were generated when they were incubated with white-egg. Thus, DNA crown cells are artificial cells which the outside of the membrane were covered with DNA and could replicated within egg white. These DNA crown cells were formed as follows: first, Sph-DNA mixtures aggregates with adenosine-lipids. From aggregates, DNA crown cells were constructed with lipids [9,10]. Thus, the mechanism on the formation of DNA crown cells became clear. Un-limited DNA crown cells could be theoretically prepared. and using the methods, the several kinds of DNA crown cells have been synthesized [11-14]. The detail on DNA crown cells (discovery, biological preparation, synthesis and the mechanism of the formation, and so on) have been described [6,15-18]. On the other hand, it is not clear whether such DNA crown cells can contribute in the applied fields, such as biomedicine or bio-industry. Here, to research these problems, it was tried to prepare the DNA crown cells (named DNA (S. griseus) crown cells) using DNA from Streptomyces griseus which produce several kinds of antibiotic and .it was examined whether such DNA crown cells were associated with the production of antibiotic. In present experiments, it was described that DNA (S. griseus) crown cells could be prepared with the method described previously and the antibiotic were produced in the co-cultures of yeast (beer).
Materials and Methods
Materials
The following materials were used:
I. The materials to prepare DNA crown cells: Sph (Sigma, USA), DNA (extracted from Streptomyces griseus), adenosine (Sigma, USA and Wako, Japan), monolaurin (Tokyo Kasei, Japan) Edible white leghorn eggs punched from a market. A-M compound (synthesized with the mixtures of adenosine and monoraulin) [10].
II. The materials to prepare the sample in co-cultures: Yeast: Dry Ale Yeast (Safale S-04) (Formentiis Bergy). Medium (Molt): Black Rock PISENER (New Zeiland) (These materials were involved in Handmade-beer kit (AUBERCRAFT, Okazaki, Japan) and was prepared as indicated in explanation.
III. The materials to test antibiotic
a) Medium: Potato Dextrose Agar (kyodo-nyugyo, Tokyou, Japan).
b) Tested bacteria: Dry Bacillus subutiis natto (Daikokuya, Nagoya, Japan)
These bacteria were suspended in distilled water (10mg/ml).
Methods
Preparation of DNA (S. griseus) crown cells: The generation of artificial cells using Sph-DNA-A-M was tested as described above [10]. Briefly, 90 μl of Sph (10 mM) and 40μl of DNA (1.7μg/μl) were mixed and the mixture was then heated. A-M compound (50 μl) was added, then the mixture was injected into the white (albumin) of an egg. After injection, eggs were incubated at 37℃ for 7days.
Observation of St DNA (S.grisses) Crown Cells: After incubation at 37 °C for 7 days, 1–2ml of egg white potentially containing artificial cells was transferred to Dulbecco’ modified Eagles’ Medium containing 10% bovine serum (DMEM) and incubated at 37 °C for 2 days. A drop of precipitate in culture medium was placed on a glass slide after the addition of ethidium bromide solution and observed using phase contrast microscopy and fluorescence microscopy.
Maintenance and storage of DNA Crown Cells: DNA crown cells were transplanted by inoculating a sample of egg white (0.5ml) containing the cells into fresh egg white every about 1 month. The part of the white albumin was storage at a storage at temperature of 4℃.
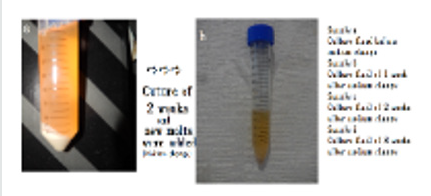
Preparation of sample (beer) tested antibiotic: Dry yeast (3g) and white egg (15ml) which contain DNA (Str. griseus) crown cells in 3 generations were mixed and incubated for 5 hours at 37℃. Then, 30ml of beer molts were added and incubated for 2 weeks in room temperature. After 2 weeks, old medium was deposit. Then, 30ml of new molts were added and continue to incubate for room temperature. In each time, medium was mixed and about 5~6 ml was removed for testing.
i. Sample a: culture fluids after 2 weeks (before medium change)
ii. Sample b: culture fluid in 1 week after medium change
iii. Sample c: culture fluid in 2 weeks after medium change
iv. Sample d: culture fluid in 3 weeks after medium change
Preparation of plate to assay antibiotic: Antibiotic assay was carried out with agar well methods Tested bacteria (5ml) were added to agar (200ml) and mixed. Then, about 15ml of agar were pored to a dish. After fixing, a well of about 2cm in diameter each dish. were prepared. Then, tested fluid (400μl) were pored to each dish and incubated for 18 hours at 37℃. After incubation, inhibition zone was observed.
TLC study on antibiotic: To prepare the samples of TLC, Acetone was added to 50ml of sample d. Then, ethyl acetate was added to the fluid and concentrated in 5ml. Extracted sample was dried and resolved in acetone (100μl) The 20μl was applied on TLC plate with Streputomaysin and Chromomysin as a control. Then, the plates were developed with chloroform/methanol (80:20) and then detected with UV 365 and 254.
Results and Discussion
Preparation of DNA (S. griseus) Crown Cells
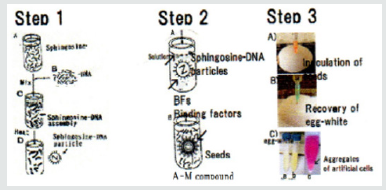
Figure 1: Summarized procedures to prepare and synthesize generated artificial cells (DNA crown cells) [7,12].
Step 1: Sph is added to DNA. Sph-DNA particles are obtained following heating of the mixture.
Step 2: To prepare artificial cells (DNA crown cells), a binding factor is added to the Sph-DNA particles. To synthesize DNA crown cells, adenosine-monolaurin (A-M) is added to the Sph-DNA mixture (Sph-DNA-A-M), then monolaurin is added.
Step 3: Sph-DNA-A-M mixtures are injected into egg white. After incubation for 7 days, cells are recovered from the egg white.
The methods on preparation of DNA crown cells (artificial cells) have been established using several kinds of DNA [10,12,13]. Summarized procedures to prepare artificial cells (DNA Crown Cells) have been shown in previous report [7], however, the methods were again shown in Figure 1
a) Step1: Sph and DNA were mixed, and heated, Sph-DNA particles were obtained.
b) Step 2: Binding factor was added to Sph-DNA particles.
Here, A-M compounds as a binding factor were added to Shp- DNA particles.
Sph-DNA-A-M mixtures were formed. After the addition of monolarin, DNA crown cells were formed without egg white.
c) Step 3: Multiplication of DNA crown cells within egg white
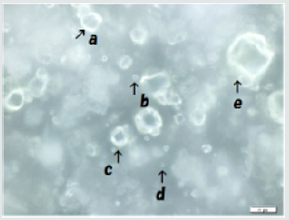
DNA crown cells could be multiplied within egg white: In use of DNA from Streptomyces griseus, Sph was mixed with DNA (S. griseus) and heated. Then, A-M compounds were added to Sph-DNA mixtures. the mixtures were incubated in egg-white. After 7days of incubation, the egg-white was removed and then incubated within egg white of new egg. After the transplantation of 3~5 times (3~5 generations), egg white were removed and DNA (S. griseus) crown cells were recovered from egg white with the incubation of D-MEM. DNA (S. griseus) crown cells in three generations are shown in Figure 2. Many cells of various sizes (approximately 2~30μm) were observed in rings like shape (Figure 2). Typical cells were shown in Figure 3. Ring shaped cells were observed under phase contrast microscopy (Figure 3a), Russell light was observed in the cells under fluorescence microscopy (Figure 3b), suggesting that they contain DNA. These findings indicate that DNA (S. griseus) crown cells were formed from the Sph-DNA-A-M mixtures.
Figure 2: Preparation of generated DNA (S. griseus) crown cells. Sph was added to DNA. After heating, A-M was added to the Sph-DNA mixture, followed by the addition of monolaurin. The mixtures contained DNA crown cells were incubated in egg white. The egg white was transferred into a new egg every 7 days. An aliquot of third generation egg white (2ml) was cultivated in Dulbecco Modified Eagle’s Medium containing 10% bovine serum at 37°C for 2 days. The precipitates were observed using phase contrast microscopy and fluorescence microscopy. Various-sized cells were observed under phase contrast microscopy (Figure 2). a and c is approximately 20 μm b and d is approximately 4 μm. e is approximately 30μm.
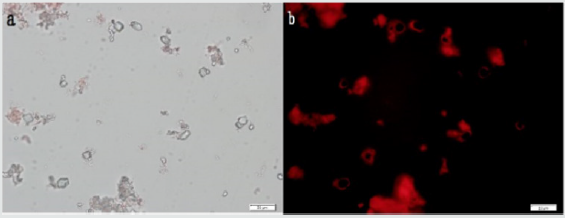
Figure 3: Typical DNA (S. grises) crown cells were observed using phase contrast microscopy (Figure 3a). Russert light was observed on the surfaces of the cells using fluorescence microscopy, indicating the presence of DNA on the surfaces of the cells (Figure 3b). Figures 3a and 3b show the same field of view. Scale bar is 20mm.
Obvious characteristics of DNA (S. griseus) crown cells are
a) The structure is a large loop of DNA
b) The cells can replicate in egg white.
These observations are similar to those of other DNA crown cells which were described previously.
Antibiotic production in co-cultures
Update, it was described that many kinds of DNA crown cells could be prepared. However, it is unclear whether these DNA crown cells could be contributed in applied fields, especially, bio-industry. Here, to research these problems, first, it was tried to prepare DNA crown cells (named DNA (S. griseus) crown cells) using DNA of Streputomysec griseus which produced Chromomycin or Strepuomycine. The second, it was examined whether antibiotic was produced with the co-cultures of yeast and egg white contained DNA (S. griseus) crown cells. The methods to obtain sample were shown in Figure 4 .
Figure 4: Preparation of sample tested antibiotic Yeast was mixed with egg white which contain DNA crown cells. The mixtures were incubated at 37 oc for 5 hours. Then, new molts were added (A) and incubated for 2 weeks at room temperature. After 2 weeks, old medium was removed, and new molts were added and continue to incubate in room. At each time point, about 5-6ml of medium was removed for testing (B).
i. Procedure 1: Yeast was mixed with egg white which contained DNA (S. griseus) crown cells. Then, the mixtures were incubated for 5 hours at 37℃.
ii. Procedures 2: After incubation, the upper fluid in the mixtures were removed and new molt was added to the mixtures. Then, the mixtures were incubated for 14 days at room temperatures (approximately 20℃).
iii. Procedures 3: After 2 weeks of incubation, the old molt was removed, and new molt was added (medium change). The part of the old molt was collected and used as a sample (a).
The molt which was carried out the medium change were further incubated at room temperatures. Then,
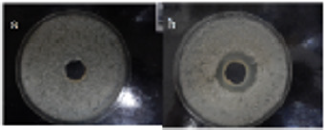
After 1 weeks of incubation, the upper fluid was collected and used as a sample (b). After 2 weeks of incubation, the upper fluid was collected and used as a sample (c). After 3 weeks of incubation, the upper fluid was collected and used as a sample (d). Each sample was collected three times within a 2~3 days and antibiotic assay was tested. Here, the prove on whether antibiotic was produced were based on whether the clear zone was observed. No antibiotic was found in sample a (Fluid after 2 weeks incubation). Also, no antibiotic was founded in sample b and sample c. The result in sample b was shown in Figure 5a as a negative example. No clear zone was founded, indicating that antibiotic was not contained in the sample. Samplings in sample d were carried out three times and antibiotic were found in a sampling of one time. The clear zone was observed as shown in Figure 5b, indicating that antibiotic was contained in sample d. Thus, the results indicated that antibiotic was produced with the co-cultures of yeast and egg white which contained DNA (S. griseus) crown cells.
Figure 5: Antibiotic production Tested bacteria (Bacillus subcilis (natto) were added to agar. Then, it was pored to a dish. After fixing, a well was prepared. Tested sample (400μl) was pored and incubated for 18 hours at 37oc. After incubation, inhibition zone was observed Inhibition zone was not observed in sample a (Figure 5a), suggesting that antibiotic was not produced. Inhibition zone was not observed in sample b and c, respectively (Data not shown). Inhibition zone was observed in sample d (Figure 5b), suggesting that antibiotic was produced.
TLC study on antibiotic
Sample (d) was treated with acetone and concentrated. Bactericidal ability did not lost with the treatments (data not shown). The sample was spotted on the TLC plates. Then, the plates were developed. Band of samples were not fond in the RF value of Chromomycin and Streptomycin, respectively. (Data was not shown). The facts showed that antibiotic which was produced with the co-cultures differ from Chromomycin or Streptomycin. It is well known that DNA from bacteria is transferred into eukaryotes in trans-king dome conjugation) [19,20]. Therefore, I expected that the Chromomycin or Streptomycin were produced with the transfer of DNA crown cells (or their DNA) to yeast. However, in present experiments, it is not possible to clear whether the transfer occur. Therefore, now, there are several unresolved problems on the mechanisms of antibiotic production. Especially, why dose it takes for long time (over 5 weeks) until antibiotic was produced, During the time, what happen between DNA crown cells and yeast. Also, antibiotic production was observed in one case. why antibiotic production was not observed in all cases. Thus, it is unclear whether DNA crown cells are directly associated with the production of antibiotic or egg white-components are associated. It is under studying to resolve these problems including the identification of antibiotic, its’ biological functions (anticancer, antibacterial spectrum and so on) of the antibiotic and weather DNA crown cells or the components were incorporated into yeast, moreover, yeast which produce antibiotic could be cloning. Thus, though it is studying on the mechanisms on the antibiotic production, it may be told that antibiotic of new type or potential antibiotic were produced with yeast in the co-cultures. On the other hand, it may be good results for apply these cells, or their egg white in the fields of bio-industry, because the procedures is very easy, and then beer containing antibiotic were produced in large scale. Namely, the production of antibiotic or functional beer with antibiotic was produced with present co-culture systems. If obtained antibiotic was effective and beer were proved in the safety, it may be actually used in the fields of medicine or healthy drink. Moreover, the present experiments expected to develop in the fields of fermented industry using Yeast.
Conclusion
Here, first, it was described that generating DNA (S. griceus) crown cells could be prepared. Second, it was demonstrated that antibiotic was produced in the co-cultures of white egg which contained DNA crown cells and yeast (beer). Now, it is unclear whether DNA crown cells are directly associated antibiotic production. However, it is expected that the production of antibiotic or beer with antibiotic was developed. The present experiments showed a possibility which it may be applied in the fields of bio (fermented) industry using yeast.
Acknowledgement
I would like to thank M. Hayakawa and H. Yamamura (Yamanashi University) for the supply of Streptomyces grises and for useful discussion. Also, I would like to thank I. Monna (Rizo Inc.) for assistance in extracting samples for adenosine analysis and for useful discussions, S. Higuchi (Saitama Industrial Technology Center Northern Laboratory, Japan) for assistance with analysis of the TLC and for useful discussions, and Tokyo Metropolitan Research Industrial Technology Institute for the use of laboratory space and fluorescence microscopes.
References
- Zhang Y, Ruder WC, Leduc PR (2008) Artificial cells; Building bioinspired systems using small-scale biology, Trends Biotechnol 26(1): 14-20.
- Liu YJ, Hansen GP, Venancio Marques A, Akiyoshi K (2013) Cell-free preparation of functional and triggerable giant proteoliposomes. Chembiochem 14(17): 2243-2247.
- Kuruma Y, Stano P, Ueda T, Luisi PL (2009) A synthetic biology approach to the construction of membrane proteins in semi-synthetic minimal cells. Biochim Biophys Acta 1788(2): 567-574.
- Noireaux V, Maeda YT, Libchaber A (2011) Development of an artificial cell from self-organization to computation and self-reproduction. Proc Natl Acad Sci USA 108(9): 3473-3480.
- Inooka S (2012) Preparation and cultivation of artificial cells. App Cell Biol 25: 13-18.
- Inooka S (2016) Investigation of the chemical composition of artificial cell seeds; Sphingoisne-DNA bound components from extract of the meat from adult ascidians. International Journal of Current Research in Life Science 5: 534-540.
- Inooka S (2016) Preparation of Artificial Cells Using Eggs with Sphingosine-DNA. J Chem Eng Process Technol l7: 277.
- Inooka S (2016) Aggregation of sphingosine-DNA and cell construction using components from egg white. Integrative Molecular Medicine 3(6): 1-5.
- Inooka S (2017) DNA crown Cells. Synethesis and Self-Replication. International Journal of Biotechnology and Bioengineering 3(1): 30-32.
- Inooka S (2017) Systematic Preparation of Artificial Cells (DNA Crown Cells). J Chem Eng Process Technol 8: 327.
- Inooka S (2017) Systematic Preparation of Bovine Meat DNA Crown Cells App. Cell Biol, Japan 30: 13.
- Inooka S (2018) Systematic Preparation of DNA (Akoya pearl oyster) Crown Cells App. Cell Biol, Japan 31: 21-34.
- Inooka S (2013) Preparation of Artificial Cells for Yogurt Production App. Cell Biol 26: 13-17.
- Inooka S (2014) Preparation of Artificial Human Placental Cells App. Cell Biol 27: 43-49.
- Inooka S (2017) Biotechnical and Systematic Preparation of Artificial Cells (DNA Crown Cells). Global Journal of Research in Engineering 17(1).
- Inooka S (2009) Cysto-organisms (cell-originated cultivable particles) with sphingosine-DNA. Comm App Cell Biol 7 (1).
- Inooka S (2014) Theory of cyto-organisms generation (sequel). The track of the dawn of self-replicating artificial cells (in Japanese). Daigaku kyouiku Press. Okayama, Japan.
- Inooka S (2018) Theory of cyto-organisms generation. The artificial cells were formed with eggs. The discovery and synthesis of DNA Crown Cells. (in Japanese). Daigaku Kyoiku Press. Okayama. Japan.
- Bogumil J Karas, Jelena Jablanovic, Lijie Sun, Li Ma, GM Goldgof, et al. (2013) Direct transfer of whole genomes from bacteria to yeast. Nat Methods. 10 (5): 410-412.
- Filip Husnik and John p. Mccutcheon (2018) Functional horizontal gene transfer from bacteria to eukaryote. Nature Reviews: Microbiology 16: 67-79.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...