
Lupine Publishers Group
Lupine Publishers
Menu
Research Article(ISSN: 2641-6875) 
Cell Proliferation from the Assembly of Synthetic DNA (E. Coli) Crown Cells with Stimulation by Monolaurin Twice Volume 2 - Issue 5
Shoshi Inooka*
- Japan Association of science specialists, Japan
Received: June 22, 2022; Published: June 29, 2022
*Corresponding author: Shoshi Inooka, Japan Association of science specialists, Japan
DOI: 10.32474/CTBM.2022.02.000149
Abstract
DNA crown cells (artificial cells), in which the outside of the membrane is covered with DNA, can be readily synthesized in vitro using sphingosine (Sph)-DNA-adenosine mixtures to produce synthetic DNA crown cells. As these DNA crown cells can proliferate within egg whites, many kinds of synthetic DNA crown cells and DNA crown cells can be prepared using DNA from a variety of donors. Though the methods for synthesis are well established, how synthetic DNA crown cells are able to proliferate in egg whites remain unclear. Previous studies have demonstrated that assemblies of synthetic DNA (E. coli) crown cells were formed in response to the inorganic substances contained in egg white, and that modified synthetic DNA crown cells were produced within such assemblies. Moreover, the cells in these assemblies were not released after the addition of monolaurin. Rather, the assembly formed crystal-like substances and the cells disappeared. In the present study, it was examined whether cells were regenerated with two monolaurin treatments. Here, it was described that cells were produced from the assembly of synthetic DNA (E. coli) crown cells which form crystal-like substances with stimulation by monolaurin twice. The cell proliferation experiments were carried out as follows:
a) Synthetic DNA (E. coli) crown cells were incubated for 18 hours at 37℃ and these cells formed assemblies.
b) Monolaurin was added to the assemblies and the mixtures were incubated for 36 hours at 37℃.
c) The assemblies then transformed into crystal-like substances.
d) Monolaurin was added to these reconstructed synthetic DNA (E. coli) crown cells and the mixtures were incubated for 20–40 minutes at 37℃, which is when cell proliferation was observed.
Keywords: Synthetic DNA Crown Cells; Assembly; Sphingosine-DNA; Cell Proliferation
Introduction
Approaches for generating self-replicating artificial cells, which were cells covered with DNA and referred to as DNA crown cells, have been reported previously [1-3]. DNA crown cells can be prepared using both biotechnology and synthetically [4]. Synthetic DNA crown cells can be prepared with sphingosine (Sph)-DNA and adenosine -monolaurin (A-M) compounds, and DNA crown cells can be generated by incubating synthetic DNA crown cells in egg white. In theory, unlimited numbers of synthetic DNA crown cells and DNA crown cells can be prepared [5-8]. While the methods for producing DNA crown cells are well established, how these synthetic DNA crown cells proliferate within egg white has not yet been clarified. In a previous study [9], the assemblies of synthetic DNA (E.coli) crown cells were produced using commercial salt, NaCl, KCl, MgCl2, and FeCl3 solutions. In addition, many synthetic DNA (E. coli) crown cells that were produced spontaneously were retained within the formed assembly. Such assemblies formed crystal- like substances after the addition of monolaurin, and the cells within the assembly disappeared [10]. The present study examined whether cells can be regenerated from the assembly of synthetic DNA (E. coli) crown cells, which changed to crystal-like substances. In the present study, it was demonstrated that cells could be regenerated from these assemblies with the addition of monolaurin. The protocol used to regenerate these cells is described below.
Materials and Methods
Materials
The following materials were used: Sph (Tokyo Kasei, Japan); DNA (E. coli B1 strain, Sigma-Aldrich, USA); adenosine (Sigma- Aldrich and Wako, Japan); and monolaurin A-M (Tokyo Kasei), a compound synthesized from a mixture of adenosine and monolaurin) [11]. Monolaurin solutions were prepared to a concentration of 0.1M in distilled water.
Methods
Preparation 0f DNA Crown Cells
The generation of artificial cells using Sph-DNA-A-M was performed as described previously [11]. Briefly, 180 μL of Sph (10 mM) and 90 μL of DNA (1.7μg/μL) were combined, and the mixture was heated twice. A-M solution (100 μL) was added, and the mixture was then incubated at 37 ºC for 15 minutes. Next, 30 μL of monolaurin solution was added and the mixture was incubated at 37 ºC for another 5 minutes. The resulting suspension was used as the synthetic DNA (E. coli) crown cells.
Test for Cell Proliferation
a) 25 μL of synthetic DNA crown cells was incubated for 18 hours at 37℃.
b) 25 μL of monolaurin was added to synthetic DNA crown cells
c) The mixture was incubated for 36 hours at 37 ºC
d) Then, 25 μL of monolaurin was added to the mixtures and incubated for 20-40 minutes at 37℃.
e) Each sample (steps 3 and 4) was observed under a light microscope.
Test for Cell Proliferation
A drop of sample was placed on a slide glass and covered with a cover glass. The slides were then observed under a light microscope.
Results and Discussion
Microscopic Appearance of Monolaurin-Treated Synthetic DNA (E. Coli) Crown Cells Before Addition of Monolaurin Twice
Synthetic DNA (E. coli) crown cells were incubated for 18 hours, then in the presence of monolaurin for 36 hours (Figure 1). Numerous crystal-like substances were observed (Figure 1, arrows a and c). Adhesive substances other than crystal-like substances were also observed (arrow d). Objects that were not transformed to crystal- like substances was observed (arrow b). A free synthetic DNA crown cell (approximately 3–5 μm) was also observed (Figure 1, arrow e). No cell proliferation was observed in Figure 1. As reported previously, the assembly of synthetic DNA (E. coli) crown cells that formed spontaneously changed to crystal-like substances after the addition of monolaurin [11]. Figure 1 shows that monolaurin- treated synthetic DNA (E. coli) crown cells formed a crystal-like substance.
Figure 1: Microscopic appearance of monolaurin-treated synthetic DNA (E. coli) crown cells before stimulation with monolaurin twice. Synthetic DNA (E.coli) crown cells were incubated for 18 hours, then in the presence of monolaurin for 36 hours at 37oC. Numerous crystal-like substances were observed (Figure 1, arrow d). Clear crystal-like substances were observed (Figure 1, arrow d). Mucoid-type aggregates were observed (Figure 1, arrow d). Objects that may not have been transformed into crystallike substances were observed (Figure 1, arrow b). A free synthetic DNA crown cell (approximately 3–5 μm) was observed (Figure 1, arrow e).

Cell proliferation
Figure 2: Microscopic appearance of synthetic DNA (E. coli) crown cells that were incubated with monolaurin for 36 hours at 37oC and 20–40 minutes at 37oC. Numerous cell-like objects were observed within the assembly (Figure 2, arrow a). Large objects (floating objects) were observed within the assembly (Figure 2, arrow b) A free cell was observed (Figure 2, arrow c, size 10-15 μm).
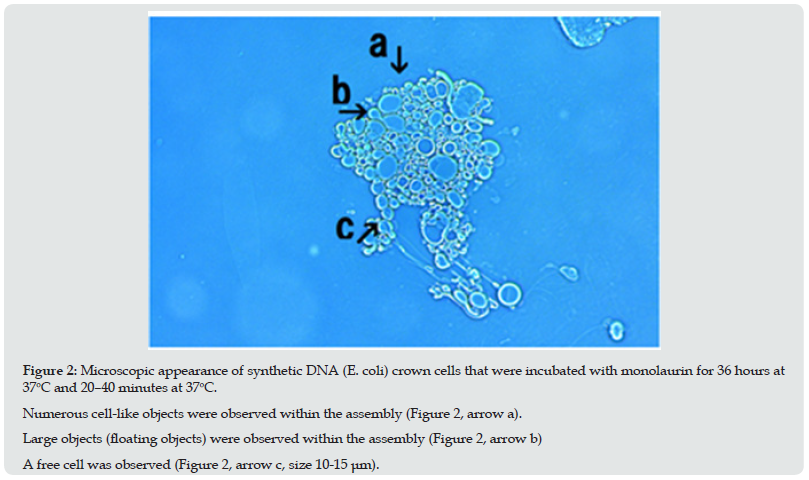
Figure 3: Microscopic appearance of synthetic DNA (E. coli) crown cells that were incubated with monolaurin for 36 hours at 37oC and 20–40 minutes at 37oC. The assembly consists of cell-like objects (Figure 3, arrow d) and floating objects (Figure 3, arrows b and c) were observed (Figure 3, arrow a). Also, an assembly consisting of cells of various sizes was observed (Figure 3, arrow e). Long tubular structures, possibly derived from the floating objects (Figure 3, arrow h) were observed (Figure 3, arrow g). A free cell (approximately 20-25 μm) was observed (Figure 3, arrow f).

Figure 4: Microscopic appearance of synthetic DNA (E .coli) crown cells that were incubated with monolaurin for 36 hours at 37oC and for 20–40 minutes at 37oC. The assembly, which consisted of floating objects of various shapes and sizes (Figure 4, arrows c, b and d) and cells (approximately 15–20 μm) (Figure 4, arrow e) were observed (Figure 4, arrow a). An assembly consisting of cells was observed (Figure 4, arrow f). A tube which was derived from a floating object was observed (Figure 4, arrow g).
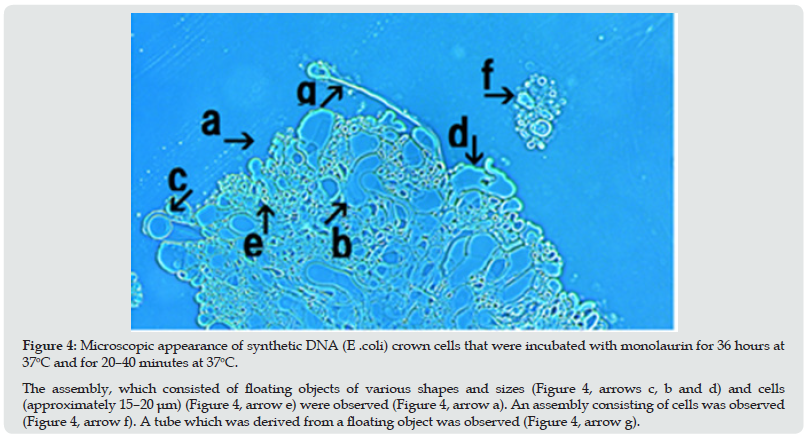
Figure 5: Microscopic appearance of synthetic DNA (E. coli) crown cells that were incubated with monolaurin for 36 hours at 37oC and 20–40 minutes at 37oC. Microscopic appearance of proliferating cells that may have been derived from one assembly (Figure 5, arrow d). In addition, the following structures are shown in the figure: A population of many separated cells (Figure 5, arrow d); a large ring-like structure composed of two rings (Figure 5, arrow d); a long tubular structure (Figure 5, arrow d); a balloon-like structure (Figure 5, arrow d), incomplete ring-like structures (Figure 5, arrows d); objects that were not transformed to crystal-like substances (Figure 5, arrow d); a cell population consisting of a few cells (Figure 5, arrow d); and a free cell (approximately 20–25 μm) (Figure 5, arrow d).to crystal-like substances (Figure 5, arrow d); a cell population consisting of a few cells (Figure 5, arrow d); and a free cell (approximately 20–25 μm) (Figure 5, arrow d).

Monolaurin was added to synthetic DNA (E. coli) crown cells which were incubated for 18 hours at 37℃and the mixture was incubated for 36 hours at 37℃. followed by incubation at 37℃ for 20–40 minutes. Cell proliferation was observed during incubation as shown in Figures 2-5. Numerous cell-like objects were observed within the assembly (Figure 2, arrow a). Among these, large objects which may begin to move were observed (Figure 2, arrow b). For convenience, these objects are referred to here as floating objects. A free cell was also observed (arrow c, size 10–15 μm). Figure 2 appears to show the microscopic appearance of the assembly before cell proliferation begins. The assembly (Figure 3, arrow a) which consists of synthetic DNA crown cells like objects (Figure 3, arrow d) and floating objects (Figure 3 arrow b) were observed. Also, an assembly consisting of cells of various sizes was observed (Figure 3, arrow e). A long tube, which may be derived from the floating objects shown in Figure 3 (arrow h), was observed (Figure3, arrow g).
A free cell (approximately 20–30 μm) was also observed (Figure 3 arrow f). The assembly shown in Figure 4 (arrow a), consisting of floating objects of different shapes and sizes (Figure 4, arrows b, c and d) and a cell (approximately 15–20 μm), were also observed within the assembly (Figure 4 arrow e). An assembly consisting of cells of different shapes and sizes was observed (Figure 4, arrow f). A tube, which was derived from the floating object was observed (Figure 4, arrow g).Figure 5 (arrow a) shows the microscopic appearance of proliferating cells, which may be derived from an assembly of synthetic DNA crown cells. A population of free cells was observed (Figure 5 arrow b). A large, double ringed, ring-like object was observed (Figure 5 arrow c).Objects similar in appearance to incomplete rings of various sizes were observed (Figure 5 arrows f, g, and i). A population of cells consisting of several cells was observed (Figure 5, arrow h). Tube-like objects were observed (Figure 5, arrow d), and balloon-like structure was observed (Figure 5, arrow e). Objects that may not have transformed to crystal-like substances were also observed (Figure 5, arrow k). A single cell (approximately 10–15 μm) was observed (Figure 5, arrow m). These figures, especially, Figure 5 may show the various stages in the process of cell proliferation. Based on these figures, cellular proliferation is described below. First, the assembly of synthetic DNA crown cells (Figure 1), which was mostly transformed to crystal-like substances, was regenerated to form a new assembly (Figure 2) consisting of cells and floating objects, Then, various tubular structures (Figure3 g, Figure 4 g, Figure 5 d) and balloon-like structures (Figure 5 e) that may have been derived from the floating objects (Figure3, h) formed. Cells may be formed the from the elongated tubular structures. These tubular structures consist of bunches of Sph. DNA, as described previously [11,12]. When a tubular structure became thin, it formed circular objects (Figure 5, arrows c and g). Moreover, thick circular objects (Figure 5 arrow c) may appeared to divide into thinner and developed into small and thing objects. Most of the cells (Figure 5 arrows h and i) were regenerated from these circular objects (Figure 5 arrow c). Another process may be responsible for regenerating new cells, that is, that the objects that spout from the elongated tubes, or fragments of elongated tubes, developed into new cells Thus, it appears that several possible explanations exist for the observed the mechanisms of cell proliferation. However, the primary aim of this study was to describe the protocol used to produce cells from the assembly, and this process can be summarized as follows:
Step 1: Synthetic DNA crown cells were cultured for 18 hours at 37℃, and these cells formed assemblies.
Step 2: Monolaurin was added to the assemblies of synthetic DNA crown cells and the mixtures were incubated for 36 hours at 37℃. The assemblies then transformed into crystal-like substances.
Step 3: Monolaurin was added to the transformed assemblies and the mixtures were incubated for 20–40 minutes at 37℃and cell regeneration occurred. Importantly, steps 2 need to be performed over 36 hours.
Many experiments were carried out on the relationships between cell proliferation and the timing of monolaurin addition to the assemblies of synthetic DNA crown cells that then transformed into crystal-like substances. Finally, it was found that assemblies needed to be incubated with monolaurin for over 36 hours but not less than 18 hours. However, to date, the mechanisms underlying this time requirement is unclear.
Cell proliferation (Step 3) begin at 37℃ after 20–40 minutes of two additions of monolaurin stimulation.
The findings suggested that cell proliferation occurs when synthetic DNA crown cells were reconstructed three times. The first occurred when synthetic DNA crown cells were cultured and the cells were reconstructed during assembly formation. The second occurred when the assembly of synthetic DNA crown cells was cultured with monolaurin and the assembly was transformed into crystal-like substances.
The third occurred when monolaurin was added to the transformed assembly and new cells were generated. As described previously [10], most of the assembly of synthetic DNA crown cells were transformed to crystal-like substances. Therefore, cell proliferation may be limited to only those assemblies that were transformed to crystal-like substances. On the other hand, it was demonstrated that lipid-adenosine (corresponding to A-M compounds) formed Sph-DNA aggregates of two types: crystalline aggregates and mucoid aggregates [3]. Therefore, floating objects may be derived from the aggregates of the mucoid-type shown in Figure 2 (arrow d) and from which cells will be generated. On the other hand, it is clear that the proliferated and regenerated cells were regenerated synthetic DNA (E. coli) crown cells, because the materials that were used in the experiments were only, Sph, DNA (E. coli), adenosine, and monolaurin. Here, it was carried out using synthetic DNA (E.coli) crown cells [14]. However, it is important to clarify whether such cellular proliferation was observed in other DNA crown cells, i.e., in addition to DNA (E. coli) crown cells, and to develop generalized methods.
References
- Inooka S (2012) Preparation and cultivation of artificial cells. App Cell Biol 25: 13-18.
- Inooka S (2016) Preparation of Artificial Cells Using Eggs with Sphingosine-DNA. J. Chem. Eng. Process. Technol. 7(1): 277.
- Inooka S (2016) Aggregation of sphingosine-DNA and cell construction using components from egg white. Integrative Molecular Medicine 3(6): 1-5.
- Inooka S (2017) Biotechnical and Systematic Preparation of Artificial Cells. The Global Journal of Researches in Engineering 17: No1-C.
- Inooka S (2017) Systematic Preparation of Bovine Meat DNA Crown Cells App. Cell Biol. Japan 30: 13-16.
- Inooka S (2018) Systematic Preparation of DNA (Akoya pearl oyster) Crown Cells App. Cell Biol. Japan 31: 21-34.
- Inooka S (2019) Preparation of DNA (Nannochloropsis species) Crown Cells Using eggs, sphingoshin and DNA, and subsequent cell recovery App. Cell Biol Japan 32: 55-64.
- Inooka S (2014) Preparation of Artificial Human Placental Cells App. Cell Biol 27: 43-49.
- Inooka S (2022) Assemblies Formation of Synthetic DNA (E.coli) Crown Cells with Salt. “Reconstruction and Regeneration of Synthetic DNA Crown Cells” American Journal of Biomedical Science & Research 16(1).
- Inooka S (2022) The assembly in synthetic DNA crown cells with inorganic salts and the transformation of DNA crown cells to crystal like substance in the presence of monolaurin Novel Research in Science 11(2): NRS.000759.
- Inooka S(2017) Systematic Preparation of Artificial Cells (DNA Crown Cells). J. Chem. Eng. Process Technol 8: 327.
- Inooka S (2018) Theory of cyto-organisms generation –Artificial cells produced with egg; the discover and synthesis of DNA crown cells (In Japanese) Daigaku kyouiku Press, Okayama, Japan p. 3.
- Inooka S (2021) Assembly and Bacterial Growth Suppression in the Co-cultures of Synthetic DNA (Akoya Pearl Oyster) Crown Cells with Bacillus subtilis. Applied Cell Biology Japan 34: 67-84.
- Inooka S (2022) Assemblies in Synthetic DNA (Nannochloropsis Species) Crown Cells with Bacillus subtilis. Acta Scientific Microbiology 5(1).

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...





