
Lupine Publishers Group
Lupine Publishers
Menu
Mini Review(ISSN: 2641-6875) 
An Overview on the Roles of Bacterial Small RNAs in Regulatory Networks Volume 1 - Issue 5
Melih AŞAN1, Mustafa AKÇELİK2 and Nefise AKÇELİK1*
- 1Biotechnology Institute, Ankara University, Ankara, Turkey
- 2Biology Department, Faculty of Science, Ankara University, Ankara, Turkey
Received:March 13, 2020; Published: June 18, 2020
*Corresponding author: Nefise AKÇELİK, Biotechnology Institute, Ankara University, Ankara, Turkey
DOI: 10.32474/CTBM.2020.01.000123
Abstract
Bacterial small RNAs (sRNAs) are important molecules that regulate the expression of certain genes, depending on the different growth conditions of the cell and they are widely used by bacteria. sRNAs help the bacteria survive by being involved in many cellular processes such as nutrient deficiency, mobility, pH adaptation and oxidative stress. Current studies have succeeded in elucidating how sRNAs modulate the expression of different transcription factors. Thus, the integration of sRNA activity into comprehensive regulatory networks has begun to take place. Regulatory networks include regulatory circuits that have characteristic functions. In this review, the roles of some characterized sRNAs in regulatory networks and their effects on transcription factors are discussed. Furthermore, we describe specific regulatory circuits containing base pairing sRNAs and their importance in global regulation.
Keywords: Small RNA; regulatory network; transcriptional regulator
Introduction
Microorganisms have to adapt extremely quickly to various challenges, particularly environmental stress conditions and the host immune system. The adaptation of bacteria to these different environments requires constant and strict regulation of gene expression [1]. Studies on bacterial small RNAs (sRNAs) have revealed that these molecules are important regulators of various cellular networks. Besides, they have key roles in mediating responses to environmental stress, regulating virulence and hostpathogen interactions [2,3]. Although it’s still a newly studied area, the regulatory roles of bacterial sRNAs have greatly expanded our knowledge of various cellular networks.
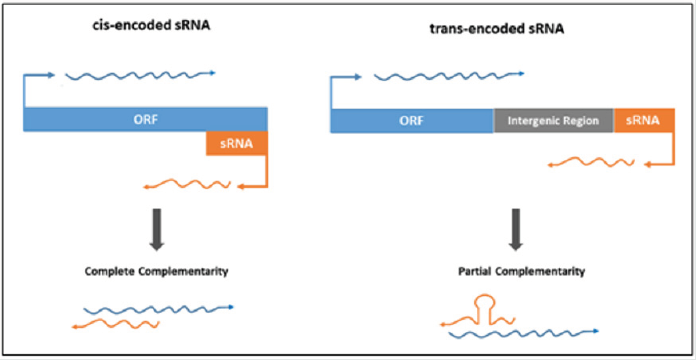
Figure 1: Difference between the mechanism of action of cis- and trans-encoded sRNAs depending on their genomic location. Orange boxes show the sRNA while blue boxes show the target mRNA. Grey box illustrates intergenic region [8].
Bacterial sRNAs are approximately 50-400 nucleotides in length
and help to modulate changes in cellular metabolism, especially
under stress conditions. As a result, the use of existing nutrients
is optimized and the possibility of bacteria survival increases [4].
While some of these regulatory RNAs can modulate the activities
of proteins by binding to them, most of the characterized sRNAs
act by performing base pairing with target mRNAs [5,6]. sRNAs
acting through base pairing are studied in two categories: (i)
trans-encoded and (ii) cis-encoded sRNAs see (Figure 1). Transencoded
sRNAs are encoded at genomic locations that are far from
the mRNAs they regulate and therefore they often share limited
complementarity with their targets [7].
Difference between the mechanism of action of cis- and transencoded
sRNAs depending on their genomic location. Orange boxes
show the sRNA while blue boxes show the target mRNA. Grey box
illustrates intergenic region [8].
Most trans-encoded sRNAs have multiple mRNA targets. In
some bacteria, base pairing between sRNAs and their targets
require the Hfq protein which is an RNA chaperone. Hfq is a protein
commonly found in bacteria that can bind to RNA, which has key
roles in controlling gene expression. Hfq affects the translation of
specific transcripts by making it easier for sRNAs to match their
target mRNAs. Unlike trans-encoded sRNAs, cis-encoded sRNAs are
transcribed on bacterial chromosomes from the opposite strand of
target genes and therefore they show great complementarity with
their targets [9,10]. In recent years, increasing characterization
studies of sRNAs have required the participation of these sRNAs
in global regulatory networks. In this review, we summarized the
various regulatory networks in which sRNAs are located and the
roles of sRNAs involved in the process. We are also focusing on the
effects of sRNA molecules interacting with mRNA targets.
Regulatory Networks Involving sRNAs
There are numerous transcriptional regulator mRNAs known
to be regulated by sRNAs. Therefore, the regulatory networks of
sRNAs may be larger than estimated. sRNAs are known to regulate
important regulatory networks such as amino acid biosynthesis,
quorum sensing system and biofilm formation. This regulation
occurs when sRNAs target mRNAs that play a role in regulatory
networks. rpoS [11], encoding the sigma factor; csgD [12] that
regulates the synthesis of curli and cellulose; luxR and aphA [13],
the quorum sensing (QS) regulators and lrp [14] involved in amino
acid biosynthesis can be given as examples to important mRNAs
that are regulated by sRNAs.
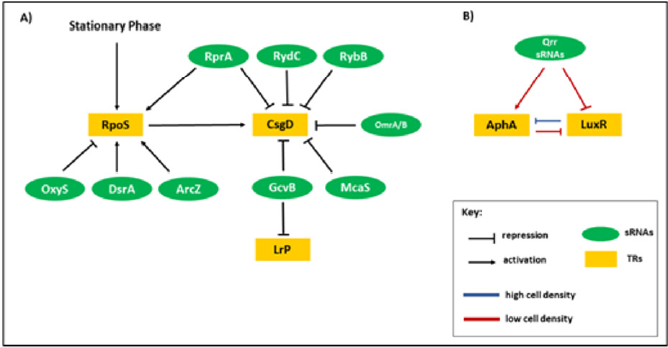
In enteric bacteria, RpoS (RNA polymerase sigma factor) protein,
a global regulator, is required to adapt to different stress conditions.
Stress stimuli, including stationary phase, nutrient deficiency, low
temperature, and osmotic shock, all cause a significant increase in
RpoS production and activity [15]. In addition, these different stress
conditions can lead to activation of Hfq-dependent specific sRNAs;
DsrA, RprA and ArcZ, which stimulate RpoS translation. In the absence of these three Hfq-dependent sRNAs, there is a secondary
structure that hides the ribosome binding site (RBS) in the 5′ UTR of
the rpoS mRNA. Each sRNA interacts with this secondary structure
of the rpoS mRNA through a similar mechanism. Then, they open the
hairpin structure and make the ribosome binding site accessible.
Thus, translational inhibition of rpoS mRNA is eliminated and the
translation is enhanced [16]. OxyS is a sRNA with a length of about
110 nucleotides that negatively regulates the translation of rpoS
mRNA. Under the oxidative stress conditions, OxyS is thought to
suppress the translation of rpoS mRNA by sequestering Hfq instead
of binding to rpoS mRNA [17].
CsgD, the main regulator of biofilm formation, is a transcription
factor that activates the synthesis of curli fimbria and extracellular
polysaccharides in Escherichia coli and Salmonella Typhimurium.
CsgD acts as a transition between planktonic and biofilm life forms
by coordinating the expression of genes involved in mobility and
adhesion [18]. The sRNAs (DsrA, ArcZ and RprA) mentioned earlier
are modulates RpoS expression and also indirectly control the
production of main biofilm matrix components, curli fimbriae and
cellulose. In addition, sRNAs called RprA, McaS, OmrA, OmrB, GcvB,
RybB and RydC cause negative regulation of the csgD mRNA by
base pairing with the 5′ UTR. Therefore, they also suppress biofilm
formation [19].
GcvB is one of the small RNAs associated with Hfq, which is
highly conserved in Gram-negative bacteria. It provides posttranscriptional
control of genes involved in amino acid metabolism
and acid stress. GcvB is known to directly inhibit the expression
of the transcription factor Lrp, which plays a role in amino acid
biosynthesis. Also, Lrp and GcvB are thought to function together in
a mutually inhibiting regulatory circuit for controlled regulation of
cellular amino acid availability [20].
The system where bacteria communicate with each other using
extracellular signal molecules known as autoinducer is called
quorum sensing. Bacteria regulate this communication process
by secreting small signaling molecules out of the cell and sensing
molecules secreted by other bacteria [21]. Five homologous sRNAs
that can regulate the QS system have been discovered in Vibrio
harveyi and Vibrio cholerae [22]. These sRNAs have been called
quorum regulatory small RNAs (qrr sRNAs). The first identified
targets of qrr sRNAs are transcription factors, the main regulator of
the QS mechanism. These are mRNA transcripts known as hapR in
V. cholerae and luxR in V. harveyi. Ongoing studies have shown that
qrr sRNAs quickly modulate QS activities depending on cell density
[13,23].
Each of the mentioned sRNAs can not only regulate these
transcription factors but also have multiple targets. All these
results show the complexity of post-transcriptional gene regulation
and the genetic network managed by Hfq and sRNAs. Regulatory
circuits involving sRNAs that regulate the rpoS, csgD, lrp, hapR and
luxR mRNAs are shown in (Figure 2).
Figure 2: Regulatory circuits involving sRNAs. A) Regulation effects of sRNAs on RpoS, CsgD and Lrp transcriptional regulators (TRs). B) QS regulation based on qrr sRNAs. At low cell density (blue arrows), qrr sRNAs stimulate translation of AphA and repress translation of LuxR. At high cell density (yellow arrow), qrr sRNA production is stopped. Therefore, the translation of AphA is stopped and the translation of LuxR is initiated [24, 25].
At high cell density (yellow arrow), qrr sRNA production is stopped. Therefore, the translation of AphA is stopped and the translation of LuxR is initiated [24,25].
Conclusion
Bacterial sRNAs are important regulators that provide control
of gene expression under specific growth conditions. These RNA
molecules, via regulating gene expression, can help bacteria adapt
to new environmental conditions and respond to various stresses.
In such a situation, sRNAs rapidly control the expression of target
genes, increasing the chances of bacteria surviving. sRNAs can
modulate transcription, translation, and mRNA stability both
positively and negatively. sRNAs that act by base pairing directly
regulate gene expression by interacting with the target mRNAs.
This interaction can occur in untranslated region or coding
sequence to induce or repress translation of the target mRNA. Also,
in many cases, the RNA chaperone Hfq facilitates the interaction
between sRNAs and their targets. Moreover, unlike the mentioned
mechanisms, some sRNAs can bind to protein targets and sequester
their functions.
sRNAs generally have multiple targets, and ongoing
characterization studies are beginning to reveal how sRNAs are
involved in global regulatory networks. Regulatory networks
consist of regulatory circuits that have characteristic behavior
and functions. Many transcriptional regulators are known to
be regulated by sRNAs. The sRNA-mediated regulation of these
transcriptional regulators led to the construction of regulatory
networks.
Thus, here, we review examples of sRNAs that are involved in
numerous cellular processes, such as stress response, adaptation
to growth conditions, quorum sensing (QS) and biofilm formation.
Research on bacterial sRNAs has many unanswered questions.
However, as a result of increased studies in which characterizations
of sRNAs are performed, new regulatory circuits will emerge. Then
we will have a better understanding of how sRNAs are integrated
into regulatory networks and why they exist.
References
- Citartan M, Raabe C, Hoe CH, Rozhdestvensky T, Tang TH (2016) Bacterial sRNAs: Regulation in Stress. In: (eds) Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria 108-114.
- Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, et al. (2010) RNAs: Regulators of bacterial virulence. Nature Reviews Microbiology8(12): 857-866.
- Holmqvist E, Wagner EGH (2017) Impact of bacterial sRNAs in stress responses. Biochemical Society Transactions45(6): 1203-1212.
- Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell136(4): 615-628.
- Pichon C, Felden B (2007) Proteins that interact with bacterial small RNA regulators. FEMS Microbiology Reviews31(5): 614-625.
- Peer A, Margalit H (2011) Accessibility and Evolutionary Conservation Mark Bacterial Small-RNA Target-Binding Regions. Journal of Bacteriology193(7): 1690-1701.
- Thomason MK, Storz G (2010) Bacterial antisense RNAs: How many are there, and what are they doing? Annual Review of Genetics44: 167-188.
- Prasse D, Ehlers C, Backofen R, Schmitz RA (2013) Regulatory RNAs in archaea: First target identification in Methanoarchaea. Biochemical Society transactions 41(1): 344-349.
- Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nature reviews Microbiology9(8): 578-589.
- Updegrove TB, Zhang A, Storz G (2016) Hfq: the flexible RNA matchmaker. Current opinion in microbiology30: 133-138.
- Battesti A, Majdalani N, Gottesman S (2011) The RpoS-mediated general stress response in Escherichia coli. Annual Review of Microbiology 65: 189–213.
- Holmqvist E, Reimegård J, Sterk M, Grantcharova N, Römling U, et al.(2010) Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. The EMBO journal 29(11): 1840-1850.
- Rutherford ST, Kessel JC van, Shao Y, Bassler BL (2011) AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes & Development 25(4): 397-408.
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf H-J, Hinton JCD, et al. (2011) Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Molecular Microbiology 81(5): 1144-1165.
- Hirsch M, Elliott T (2005) Stationary-phase regulation of RpoS translation in Escherichia coli. Journal of Bacteriology 187(11): 7204-7213.
- Papenfort K, Vanderpool CK (2015) Target activation by regulatory RNAs in bacteria. FEMS Microbiology Reviews 39(3): 362-378.
- Updegrove TB, Wartell RM (2011) The influence of Escherichia coli Hfq mutations on RNA binding and sRNA*mRNA duplex formation in rpoS riboregulation. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1809(10): 532-540.
- Liu Z, Niu H, Wu S, Huang R (2014) CsgD regulatory network in a bacterial trait-altering biofilm formation. Emerging Microbes & Infections 3(1): e1.
- Mika F, Hengge R (2014) Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biology 11(5): 494-507.
- Modi SR, Camacho DM, Kohanski MA, Walker GC, Collins JJ, et al. (2011) Functional characterization of bacterial sRNAs using a network biology approach. Proceedings of the National Academy of Sciences of the United States of America 108(37): 15522-15527.
- Jayaraman A, Wood TK (2008) Bacterial quorum sensing: Signals, circuits, and implications for biofilms and disease. Annual Review of Biomedical Engineering 10: 145-167.
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, et al. (2004) The Small RNA Chaperone Hfq and Multiple Small RNAs Control Quorum Sensing in Vibrio harveyi and Vibrio cholerae. Cell 118(1): 69-82.
- Tu KC, Waters CM, Svenningsen SL, Bassler BL (2008) A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Molecular Microbiology 70(4): 896-907.
- Shao Y, Bassler BL (2012) Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Molecular Microbiology 83(3): 599-611.
- Chambers JR, Sauer K (2013) Small RNAs and their role in biofilm formation. Trends in microbiology 21(1): 39-49.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...





