Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4676
Short communication(ISSN: 2637-4676) 
Agronomic and Economic Performance of Bread Wheat (Triticum aestivum L.) Varieties in Response to N-Fertilizer Rate at Lemu-Bilbilo District, Southeastern Ethiopia Volume 9 - Issue 5
Khin Kyawt Yin2,3, Kanogporn Khammona1, Arweewut Yongsuwan1, Naraporn Chaomueang1, Siwaret Arikit4,5, Samart Wanchana1, Jintana Unartgam2 and Vinitchan Ruanjaichon*1
- 1National Center for Genetic Engineering and Biotechnology (BIOTEC), 113 Thailand Science Park, Pahonyothin Road, Khlong Nueng, Khlong Luang, Pathum Thani 12120, Thailand
- 2Department of Plant Pathology, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Kamphaeng Saen campus, Nakhon Pathom 73140, Thailand
- 3Thailand Graduate Institute of Science and Technology (TGIST), National Science and Technology Development Agency (NSTDA), Khlong Luang, Pathum Thani, Thailand
- 4Department of Agronomy, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University Kamphaeng Saen Campus, Nakhon Pathom 73140, Thailand
- 5Rice Science Center, Kasetsart University Kamphaeng Saen Campus, Nakhon Pathom 73140, Thailand
Received: July 26, 2021; Published: August 20, 2021
Corresponding author: Vinit chan Ruanjaichon, National Center for Genetic Engineering and Biotechnology (BIOTEC), 113 Thailand Science Park, Phenytoin Road, Khlong Nueng, Khlong Luang, Pathum Thani 12120, Thailand
DOI: 10.32474/CIACR.2021.09.000321
Background
Sweetness is an economically important eating quality trait for sweet corn breeding. The various types of sweet corn are caused by single gene mutation [1]. The brittle2 and shrunken2 mutants accumulated 20% of starch found in the corn kernels [2, 3]. Due to mutation in ADP-glucose pyro phosphorylase reveals as a key regulatory enzyme to limit the rate of starch biosynthesis pathway [4-7]. The brittle2 (bt2) mutant is classified as class 1 super sweet corn [3,8,9], having 15-30% sugar of total carbohydrate content in the kernel, while normal corn contains about 3% sugar in the kernel [9,10]. Newly, the functional marker based on shrunken2 gene (sh2) was identified and developed [11]. This SNP marker designed within the coding region in exon 1 at position 154 of the coding sequence was clearly separated the sh2-based sweet corn from waxy corn, field corn, and the sweet corn that was based on the other genes, i.e., su1, se1, and bt2. However, there is no report of SNP markers associated with bt2 in sweet corn.
In this study, we scanned for SNPs from an Axiom® Maize 600k Genotyping Array from a collection of 6 sweet corn and 9 waxy corn recombination inbred lines (RILs). The SNPs were used to evaluate linkage-disequilibrium (LD) decays and association with bt2. Additionally, SNP-based makers tightly linked to bt2 were validated to find a marker associated with super sweet kernel trait that could aid in future super-sweet corn improvement programs.
Keywords: Maize; kernel Sweetness; Brittle 2; Single Nucleotide Polymorphism (SNP)
Results
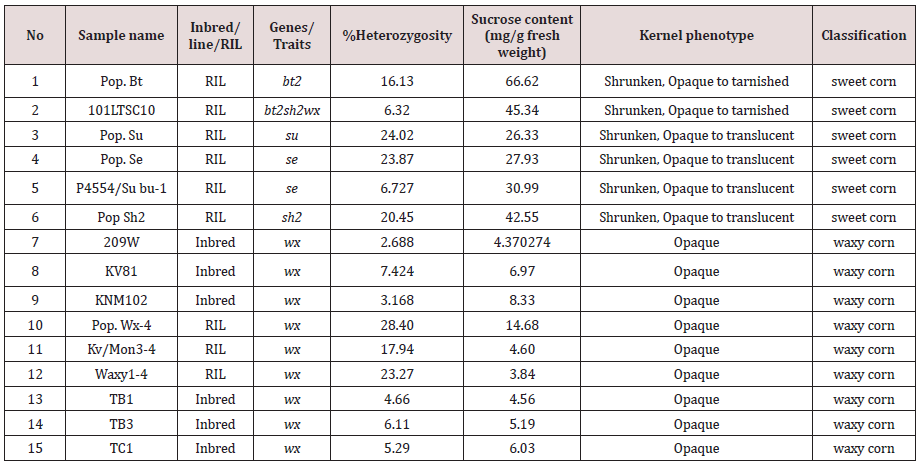
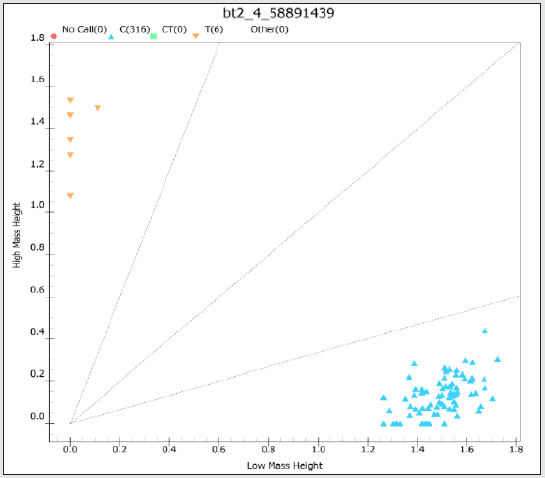
A panel of 15 maize recombinant inbred lines (RILs) were used to obtain a set of 616,201 SNP variants on the bt2 using Axiom® Maize 600K Genotyping Array (Table S1). A panel of 331,945 variants were high-quality SNPs with minor-allele frequency (MAF) > 0.05 and missing rate < 10% and were included in subsequent analysis. Chromosome-wide LD among the 15 RILs, analyzed using PLINK software [12] revealed the 210 kb LD decay at a cut-off r2 = 0.2 (Figure1). The bt2 (GRMZM2G068506) resides on chromosome 4 at position 58,954,361 to 58,960,521 bp (Maize Genome Database version2; https://www.maizegdb.org). We found that the AX-91607989 (C/T) SNP, located at 62,922 bp upstream of bt2, was significantly associated with bt2-based sweet corn (Table S2). Among the 15 RILs, 2 lines controlled by bt2 had the T allele, while the others (sh2, su, se and wx) contained the C allele. A marker was developed using Mass ARRAY® platform and validated in a diverse panel of maize composing of inbred lines, hybrid lines, and RILs (Figure2; Table S3). This marker could clearly separate the bt2- based sweet corn from waxy corn, field corn, and other types of sweet corn mutants that was based on the other genes, i.e., su1, se1, and sh2 (Figure3; Table S4).
Finally, we extended the search for gene-based SNPs linked to bt2 on chromosome 4. One SNP was validated to differentiate sweet corn from other corn types by possessing a bt2 recessive allele (T). This SNP was located upstream of the bt2 62,922 base pair. We developed a marker tightly linked to the bt2 and validated that the marker clearly distinguished the super sweetness corn from waxy corn, field corn, and the sweet corn controlled by other mutants. The identified SNPs on the bt2 could be used as selection marker for marker-assisted selection in maize-breeding programs. Moreover, the tightly linked markers identified in this study could effectively save breeders cost and valuable time from lengthy field trials, and aid in accelerating sweet corn breeding programs.
Table S2: A single marker analysis of 66 SNP AXIOM ID validated on the 15 maize recombinant inbred lines (RILs).
Table S3: The Mass Array markers Bt2_4_58891439 designed based on SNP variant (C/T) in upstream bt2.

Table S4: List of 307 inbreds and recombinant inbred lines (RILs) used formarker validation in this study.
Figure 1: Overall chromosome-wide linkage-disequilibrium (LD) decay estimated from single-nucleotide polymorphism (SNP) genotypes of 15 maize recombinant inbred lines. Each line plot represents a smoothed r2 for all marker pairs on each chromosome depending on the distance between marker pairs.

Figure 2: Physical location of the Bt2_4_58891439 SNP marker upstream of the brittle2 gene (bt2). Orange: Bt_4_58891439 SNP; gray: SNPs located in bt2 region. Sucrose content was analyzed from immature ears of each line at 24 days after pollination using the method described previously [13].
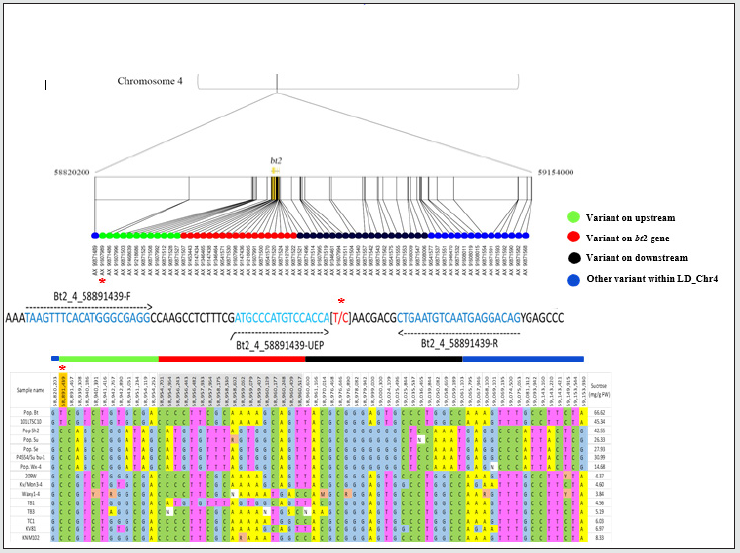
Figure 3: Allelic discrimination plot of SNP marker Bt2_4_58891439 validated in a panel of 322 maize lines as analyzed by Mass ARRAY® platform. Scatter dots with different colors showed clustering of homozygous genotype TT (yellow) and homozygous genotype CC (blue).
Funding
This research was funded by the National Science and Technology Development Agency (NSTDA), grant number P-17- 52167 and Thailand Graduate Institute of Science and Technology (TGIST), agreement number SCA-CO-2562-9848-EN and grant number TG-BT-KU-62-062M.
Data Availability Statement
Data are contained within the article and the Supplementary Materials.
Acknowledgments
The authors thank the Chai Nat Field Crops Research Center and Plant Breeding Research Center for Sustainable Agriculture, Khon Kaen University for providing the plant materials used in the study.
Conflicts of Interest:
The authors declare no conflict of interest.
References
- Douglas and Tsung, Doehlert DC, Kuo TM (1994) Gene expression in developing kernels of some endosperm mutants of maize. Plant and cell physiology 35(3): 411-418.
- Nelson OE (1980) Genetic control of polysaccharide and storage protein synthesis in the endosperms of barley, maize, and sorghum. Adv Cereal Sci Tech 3: 41–71.
- Boyer CD, Shannon JC (1983) The use of endosperm genes for sweet corn improvement. In Plant breeding reviews 139-161.
- Greene TW, Hannah LC, (1998) Enhanced stability of maize endosperm ADP-glucose pyro phosphorylase is gained through mutants that alter subunit interactions. Proc. Natl. Acad. Sci. USA 95(22): 13342-13347.
- Morell MK, Bloom M, Knowles V, Preiss J (1987) Subunit structure of spinach leaf ADP glucose pyro phosphorylase. Plant Physiol 85(1): 182-187.
- Sanwall GG, Greenberg E, Hardie J, Cameron EC, Preiss J (1968) Regulation of starch biosynthesis in plant leaves: activation and inhibition of ADP-glucose pyro phosphorylase. Plant Physiol 43(3): 417-427.
- Feng ZL, Liu J, Fu FL, Li WC (2008) Molecular Mechanism of Sweet and Waxy in Maize. International Journal of Plant Breeding and Genetics, 2: 93-100.
- Tracy WF (1997) History, genetics, and breeding of super sweet (shrunken2) sweet corn. Plant breeding reviews 14: 189-236.
- Abe A, Adelegan CA (2019) Genetic variability, heritability and genetic advance in shrunken-2 super-sweet corn (Zea mays L. saccharata) populations. Journal of Plant Breeding and Crop Science 11(4): 100-105.
- Creech RG (1965) Genetic control of carbohydrate synthesis in maize Genetics 52(6): 1175-1186.
- Ruanjaichon V, Khammona K, Thunnom B, Suriharn K, Kerdsri C, et al. (2021) Identification of Gene Associated with Sweetness in Corn (Zea mays L.) by Genome-Wide Association Study (GWAS) and Development of a Functional SNP Marker for Predicting Sweet Corn. Plants 10(6):
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira, et al. (2007) PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet 81(3): 559–575.
- Simla S, Lertrat K, Suriharn B (2010) Carbohydrate Characters of Six Vegetable Waxy Corn Varieties as Affected by Harvest Time and Storage Duration. Asian J Plant Sci 9(8): 463-470.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...