Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4676
Research Article(ISSN: 2637-4676) 
Comparison of Five DNA Extraction Methods for Detection of Leaf Scald in Sugarcane Through PCR Volume 8 - Issue 1
Vanessa Duarte Dias1*, Geisiane Alves Rocha1, Erico de Campos Dianese1, Luiz Artur Mendes Bataus2 and Marcos Gomes da Cunha1
- 1Department of pathology, Federal University of Goiás, Brazil
- 2Department of Bechemistry and Molecular Biology, Federal University of Goiás, Brazil
Received: November 19, 2019; Published: December 05, 2019
Corresponding author: Vanessa Duarte Dias, Department of pathology, Federal University of Goiás, Brazil
DOI: 10.32474/CIACR.2019.08.000278
Abstract
Among the techniques used for Leaf Scald diagnosis, PCR is considered the most sensitive, however this technique has limitations
for use in large-scale analysis, particularly because of the difficulty/complexity associated with DNA extraction. Detection success
largely depends upon the host species and the protocol used for DNA extraction. Here we describe a comparison of five DNA
extraction methods and determine which one is more efficient for large scale detection of Leaf Scald through PCR. DNA extraction
was performed from:
a. Bacterial culture;
b. Bacterial culture added to the vascular fluid from sugarcane; and
c. From infected plant stalks. Of the five methods tested, three were effective for large-scale detection by PCR.
Keywords: Xanthomonas albilineans, molecular detection, DNA extraction, large scale detection
Introduction
The sugarcane cultivation in Brazil is expanding. The production
area has increased considerably in 2011, most significantly in the
states of Minas Gerais (83,100 ha), Mato Grosso do Sul (84,700 ha),
Goiás (79,110 ha) and Mato Grosso (13 040 ha) Conab–Companhia
Nacional de Abastecimento [1]. In these states, besides increasing
the crop area, new processing plants began operating in the last
cropping season. Even with those investments, the domestic
production by the sugarcane industry in 2011/2012 was 8.4%
lower than in previous years. This decrease was due to a number
of factors. Among them, sanitary problems are remarkable, with
increased incidence of sugarcane diseases. The producers have
increased the number of cultivated areas to minimize reduction
in productivity. The expansion resulted in the multiplication of
sugarcane grown areas with no control or treatment of diseases.
According to Gatti, sugarcane diseases are very serious; since they
cause cultivar degeneration, requiring periodic replacement of
crops. Leaf scald is one of the most important worldwide diseases
of sugarcane Birch [2]. Xanthomonas albilineans, the causal agent
of Leaf Scald, has great destructive potential, capable of causing
stalk malformation and tillers death, with subsequent reduction
in sucrose content and yield losses. When the disease is present in
extremely susceptible cultivars, it may cause losses of up to 100%
Tokeshi and Rago [3].
In most cases the symptoms of this disease are not visually
noticeable, particularly due to latent infections. Thus, bacteria
multiply and spread without being detected, especially in the
beginning of the plant cycle Comstock and Irey [4]. Besides that,
symptoms that occur in a more visible way may show variations,
which may happen due to factors related to climate and plant
nutrition Tokeshi and Rago [3]. Those variations of symptoms make
the diagnosis even more difficult, requiring the use of more precise
techniques of detection, such as serological and/or molecular
methods Pan et al. [5]; Tokeshi and Rago [3]. The detection of Leaf
scald by serological techniques is effective when the bacteria reach
high concentrations in the tissue; by this time, the production is
already potentially lost and the pathogen disseminated to other
plants Honeycutt et al. [6]. PCR (Polymerase Chain Reaction)
sensibility is higher and more effective than serological techniques, detecting contaminations in samples with less than 20 UFC
of X. albilineans, whereas serological techniques as DIA (Dot
immunobinding Assay) and ELISA (Enzime Linked immunosorbent
Assay) requires populations of 106 UFC mL-1 and 105 UFC mL-1,
respectively Wang et al. [7]. PCR is the most sensitive technique
available, but it has some hindrances because it requires elaborated
protocols to extract DNA with high degree of purity, which hampers
the short time analysis of a high number of samples. Therefore, the
objectives of this work were to compare five fast DNA extraction
methods, determine the more efficient method and perform the
necessary adjustments for large scale detection of X. albilineans by
PCR.
Material and Methods
Bacteria and infected plant stalks
X. albilineans was obtained from Instituto Agronômico de Campinas (IAC) and grown in Wilbrink medium (5g of bactopeptone, 10g of sucrose, 0,5g of K2PO4.3H2O, 0,25g of Mg SO4.7H2O, 0,05g of Na2SO3 and 15g of Agar for 1 liter of culture medium). Infected sugarcane stalks were taken from experimental plots belonging to RIDESA (Rede Interuniversitária para o Desenvolvinento do Setor Sucroenergético) at Jales Machado Processing Plant, in Goianésia – GO. The infection was confirmed prior to the mailing of these samples through
Symptom Analysis
Vascular fluid of sugarcane plants
Primarily, a transversal cut was performed on the lower portions and another on the upper portion of the stalks base (between the second and third internodes). Afterwards, sap from the xylem was extracted using a low pressure compressor connected to a rubber teat cup (used in milking machines) on the hose end Gao et al. [8]. From each stalk sampled, approximately 0.3 mL of vascular fluid were obtained and transferred to 1.5 mL microtubes in which two drops of quaternary ammonia at 0.2% or chloroxidine dicluconate at 2% were added Pan et al. [5].
DNA extraction methods
Five methods of DNA extraction were tested (Table 1). All DNA extraction methods followed the original protocols, except the fourth, named M4, which was performed according to the following modifications: the initial 160 μL was centrifuged at 10,000 RPM for 10 minutes and the supernatants were discarded. The pellet was resuspended in 500 μL of Tris-EDTA (Tris 50mM and EDTA 1mM) + 2% SDS; mixed with a vortex and boiled for 5 minutes. After cooling at room temperature, 50 μL of 3M NaCl were added; samples were mixed with a vortex and 1 μL of 100% ethanol was added. Mixtures were incubated at -200C for 30 minutes followed by centrifugation at 10,000 RPM for 10 minutes at room temperature. The supernatant was discarded and the pellet resuspended in 100 μL Milli-Q water. DNA extractions were performed from three distinct sources:
a. Bacterial culture (BC)- X. albilineans cells were transferred
from Wilbrink media to 15 mL tubes containing 2.5 mL of Milli-Q
autoclaved water. The bacterial suspension was homogenized
in vortex and aliquots of 160 μL were transferred to 1,5 mL
microtubes for further use. Each DNA extraction method was made
in triplicate. To quantify the total number of bacterial cells used in
the extraction, 1 μL of this suspension was subjected to a dilution
series of 10-1 to 10-5 in a sterile saline solution (0.85% NaCl). Ten
microliters of each dilution were placed in the center of Petri dishes
with 60 mm of diameter containing Wilbrink media. Bacterial
culture added to the vascular fluid of sugarcane (BF); 10μL of the
bacterial suspension were transferred to a microtube containing
150 μL of vascular fluid from uninfected sugarcane. The vascular
fluid was obtained from five sugarcane cultivars (RB 952857, RB
925211, RB 925345, RB 867515 and RB 72454).
b. From infected plant stalks (IPS); vascular fluid from
sugarcane stalks naturally infected with X. albilineans were
extracted according to the previously described methodology.
Fifteen samples were obtained from the basal portion of these
stalks to test the five DNA extraction methods.
After the extraction, the DNA samples were quantified and the
purity was determined by reading the absorbance at 260nm and
280nm, using an Amershan Pharmacia Biotec spectrophotometer
(UV-Vis Ultraspec ® 3000 Pro). The ratio of absorbance at 260 nm
and 280 nm was used to assess the purity of DNA. Specific PCR
for X. albilineans detection. The PCRs with 20 μL of final volume
were composed by 1 μL of DNA, polymerase buffer (50 mM KCl;
10 mM Tris-HCl pH 8.3), 0.8 mM MgCl2, 0.2 mM of each dNTPs,
1.0 μM of oligonucleotide PGBL1 (5’CTTTGG GTC TGT AGC TCA GG)
and PGBL2 (5’GCC TCA AGG TCA TAT TCA GC), which amplified the
ITS regions of the 16S-23S rDNA Pan et al. [5] and one unit of Taq
DNA polymerase (Invitrogen). Amplification conditions comprised
of one cycle of denaturation at 950C for 5 minutes followed by
35 cycles at 950C for 30 seconds, annealing at 570C for thirty
seconds, extension for thirty seconds, and final extension at 720C
for ten minutes. Following the amplification, the PCR products were
analyzed by 1.5% agarose gel electrophoresis and stained with Gel
Red reagent (Invitrogen).
Results and Discussion
The efficacy of commercial kits for DNA extraction was not tested in this work, since its use in large scale analyses is economically unfeasible. These kits are generally faster than the CTAB and phenol methods; however, the cost is very high Demeke and Jenkins [9]. Real Time PCR (qPCR) is also expensive and not yet suitable for routine diagnosis tests for a large number of samples Urashima and Grachet [10]. Extraction protocols using cetil trimethyl ammonium bromide (CTAB) were widely used to extract DNA from leaves, seeds, grains, and processed food. The procedure, although efficient, is time consuming and uses hazardous chemical products, such as phenol and chloroform Demeke and Jenkins [9]. Urashima and Zavaglia [11] tested the detection of X. albilineans in vascular fluid of contaminated sugarcane by PCR using a CTAB protocol Murray and Thompson [12]. The DNA extraction step takes around 24 hours; making large scale analysis much more costly and time consuming. In this work we describe a comparison of five methods of DNA extraction and evaluate which one is more efficient for large scale detection of Leaf scald through PCR. DNA extraction was performed from: a) Bacterial culture - BC; b) Bacterial culture added to the vascular fluid from sugarcane - BF; and c) From infected plant stalks - IPS. All procedures were performed in triplicate.
Table 2: DNA quantification and purity. Concentration is expressed as mg/mL. The ratio of absorbance at 260 nm and 280 nm was used to assess the purity of DNA.
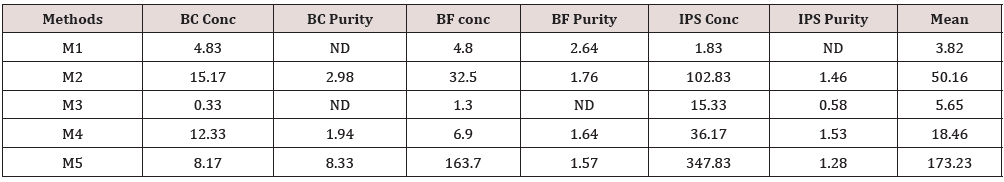
Quantification of DNA samples was estimated by spectrophotometric measurement. Table 2 shows the yield and purity of DNA extracted from each source, using the five methods. A large variation can be observed in DNA yield of each source according to each method. The concentration ranged from 0.33 to 15.17 mg/mL (BC); from 1.30 to 163.70 mg/mL (BF); and from 1.83 to 347.83 mg/mL (IPS). Methods M1 and M3 resulted in a lower amount of DNA concentration. This low concentration did not allow a precise calculation of purity levels. The M5 showed the best yield, but DNA obtained did not show an acceptable level of purity (ranged between 1.28 to 8.33). The M4 allowed the obtainment of DNA with acceptable level of purity, were the ratio was within expected values (1.53 to 1.94). The effectiveness of PCR may be affected by extraction methodology and by DNA source. Then, obtained DNA samples were used to test the influence of these variables. PCR products analysis showed that extraction methods have influence on the effectiveness of PCR. Figure 1 a) shows the influence of the extraction methods studied using BC as source of DNA. All tested methods resulted in amplifications, except for M2.
Figure 1: Products of PCR from samples originated from:
a) pure bacterial cultures;
b) vascular fluids from cultivars RB 952857, RB 925211, RB 925345, RB 867515 and RB 72454 with further addition of Xanthomonas
albilineans;
c) from infected plant stalks. L = 50 bp marker. White traces represent the replicates of each method of extraction, and the code
above each one indicates the DNA extraction methods used (M1 to M5). Arrows indicate the expected band height 288 pb).
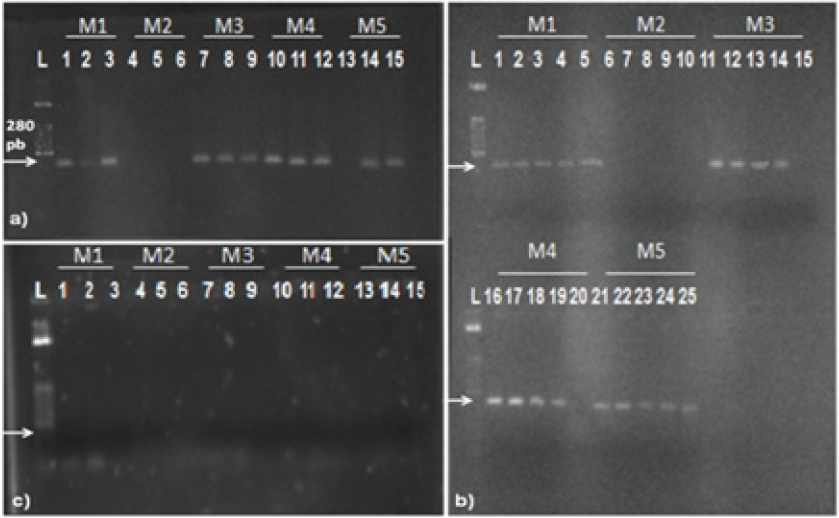
The influence of differences in vascular fluid of five cultivars of sugarcane on the efficiency of PCR can be observed in Figure 1-b. From all vascular fluid of different sugarcane cultivars tested, only one showed no reproducibility (RB 72454). Using DNA obtained by M1 and M5, PCR products were observed, but using DNA from M3 and M4 the amplification failed. Gao et al. [8] reported that vascular fluids from different cultivars of sugarcane can inhibit PCR, according to their work related with the detection of Leifsonia xyli subsp. xyli by PCR. Thirty-one cultivars of sugarcane were studied, nine were from the USA and 22 were from China. Nineteen cultivars (61.3%), tested positive by PCR, while 93.5% were positive by dot-blot enzyme immunoassay detection. This result showed the importance to evaluate the influence of nucleic acid extraction methods and the influence of different vascular fluid sources on the result of the PCR assay. Again, PCR failed for M2. M2 was effective in extracting DNA, resulting in significant quantities, however the DNA obtained by this method failed in all attempts of amplification. This result agrees with the idea that DNA extraction methodology is critical to the success of PCR Wilson et al. [13]. All assays using DNA obtained from IPS failed, no amplification products were observed Figure 1-c. The absence of amplification in all samples from IPS may be explained by the presence of PCR inhibitors in those samples. Some polysaccharides (from the plant) may act in the inhibition of the amplification of the target sequence. Many authors have reported the inhibitory action of carbohydrates on PCR Ferreira and Grattapaglia, Jaueerally-Fakin et al. [14]. Another possibility is the presence of substances in the rough extract of sugarcane with inhibitory capability of the Taq DNA polymerase, a fact that has been described by Pan et al. [5].
Figure 2: Amplification of DNA extracted from sugarcane broth with Leaf Scald;
a) amplifications from DNAs diluted in 10-1 obtained by methods M1, through M5;
b) amplifications from DNAs diluted in 10-2 obtained by methods M1, through M5;
c) amplifications from DNAs diluted in 10-3 performed by methods M1, through M5;
d) amplifications from DNAs diluted in 10-4 performed by methods M1, through M5.
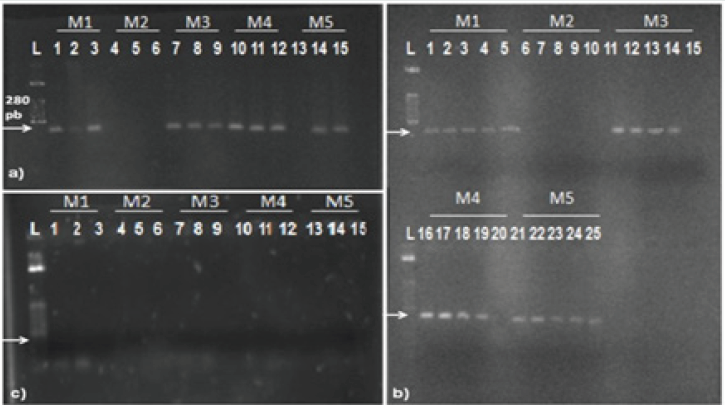
Since all assays using DNA from IPS failed, DNA obtained from this source was subjected to serial dilutions (10-1 to 10-4) to evaluate the effect on the PCR assay. PCR products analysis showed that dilutions have a strong influence on the effectiveness of PCR. PCR from IPS showed significant improvement after DNA dilution Figure 2. The dilution at 10-3 resulted in 100% of amplifications Figure 2-c and all reactions for the dilutions in question were repeated five times to test reproducibility. Results were identical for all reactions. Thereby, it was verified that PCR from diluted products of DNA, besides providing 100% of detection for all DNA methods of extraction, also achieved reproducibility. Difficulties in PCR assays using DNA from vascular fluid samples of sugarcane have been reported in the literature. Pan et al. [15] developed a protocol to detect X. albilineans by PCR using oligonucleotides ALA4 and L1; however, the protocol amplified bands of different sizes. In the following work the same authors devised highly specific oligonucleotides. PCR were carried out directly from vascular fluid of sugarcane and amplification results were not consistent Pan et al. [5]. These studies showed the influence of sugarcane´s vascular fluid on the effectiveness of PCR. Few studies have been conducted regarding PCR feasibility directly from vascular fluid over the years Comstock and Irey [4] Lopes et al. [15] Wang et al. [7]. Results from this work shows that fast methods of DNA extraction, which took approximately 2 hours for a total of 100 samples, may be as effective as more time-demanding and elaborated protocols that require around 24 hours to complete [16-20]. Therefore, large scale diagnosis of leaf scald in sugarcane may be performed through the DNA extraction techniques M2, M4 or M5, since these methods are fast, simple and do not use phenol as one of its reagents[21-23].
References
- Porto SI, Oliveira Neto AA, Sousa FOB (2012) Conab-Companhia Nacional De Abastecimento. Avaliação da safraagrícola de cana-de-açú 3ºLevantamento da safra 2012. Internet Resource.
- Birch RG (2001) Xanthomonas albilineans and the antipathogenesis approach to disease control. Molecular Plant Pathol 2:1-11.
- Tokeshi H, Rago A (2005) Doenças da Cana-de-açú In: kimati h, amoriml, rezende, JAM.; bergaminfilhoa, Camargo, LEA (eds). Manual de Fitopatologia. São Paulo, Piracicaba, pp.197-223.
- Comstock JC, Irey MS (1992) Detection of the sugarcane leaf scald pathogen Xanthomonas albilineans, using tissue blot immunoassay, ELISA, and isolation techiniques. Plant Disease 76:1033-1035.
- Pan YB, Grisham MP, Burner DM, Legendre BL, Wei Q (1999) Development of Polymerase Chain Reaction Primers Highly Specific for Xanthomonas albilneans, the causal Bacterium of sugarcane Leaf Scald disease. Plant Disease 83: 218-222.
- Honeycutt RJ, Sobral BWS, Mcclelland M (1995) tRNA intergeneric spacers reveal polymorphysms diagnostic for Xanthomonas albilineans. Microbiol 141: 3229-3239.
- Wang ZK, Comstock JC, Hatziloukas E, Shaad NW (1999) Comparasion of PCR, Bio-PCR, DIA, ELISA and isolation on semiselective medium for detection of,the causal agent of leaf scald of sugarcane. Plant Pahol 48(2): 245 -252.
- Gao SJ, Pan YB, Chen RK, Chen PH, Zhang H, et al. (2008) Quick detection of Leifsonia xyli subsp. xyli by PCR and nucleotide sequence analysis of PCR amplicons from Chinese Leifsonia xyli subsp. xyli isolates. Sugar Tech 10: 334-340.
- Demeke T, Jenkins G (2010) Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal and Bioanal Chem 396: 1977-1990.
- Urashima AS, Grachet NG (2012) Métodos de detecção de Leifsoniaxyli subsp. xyli e efeito da termoterapianabrotação das gemas de diferentesvariedades de cana-de-açú Tropical Plant Pathol 37(1): 57-64.
- Urashima AS, Zavaglia AC (2012) Comparação de doismétodosdiagnósticos de escaldadura-das-folhas (Xanthomonas albilineans) da cana-de-açú Summa Phytopathologica 38(2): 155-158.
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321-4325.
- Wilson D, Yen-Lieberman B, Reischl U, Warshawsky I, Procop GW (2004) Comparison of five methods for extraction of Legionella pneumophila from respiratory specimens. J Clin Microbiol 42: 5913–5916.
- JaueerallyFakin Y, Autrey JC, Daniels MJ, Dookun A (2002) Genetic polymorphism among Xanthomonas albilineans strains, using a single oligonucleotide primer. European J of Plant Pathol 108:121-130.
- Pan YB, Grisham M, Burner DM (1997) A Polymerase Chain Reaction Protocol for the Detection of Xanthomonas albilineans, the Causal Agent of Sugarcane Leaf Scald Disease. Plant Disease 81:189-194.
- Alsubel FN (1992) Short protocols in molecular biology. John Wiley New York, USA.
- Davis MJ, Rott P, Warmuth CJ, Chatenet M, Baudin P (1997) Intraspecific genomic variation within Xanthomonas albilineans, the sugarcane leaf scald pathogen. Phytopathol 87: 316-324.
- Gomes LH, Duarte KMR, Andrino FG, Tavares FCA (2000) A Simple Method For DNA Isolation From Xanthomonas spp. Scientia Agricola 57: 553-555.
- Lopes SA, Damann KE, Grelen LB (1998) Comparison of methods for identification of sugarcane pathogen Xanthomonas albilineans. Summa Phytopathologica 24:114-119.
- Marcuz FS, Souto ER, Vieira RA, Marrafon MA, Barboza AAL, et al. (2009) Levantamento da incidência de Leifsonia xyli subsp. xyliemplantios de cana-de-açúcar do noroeste do Paraná. Ciê Agrotec33:1935-1939.
- Ogram A, Gary SS (1987) The extraction and purification of microbial DNA from sediments. J of Microbiol Methods 7:57-66.
- Schneider S, Smith T, hansen U (2011) SCOREM: statistical consolidation of redundant expression measures. Nucleic Acids Research 40(6): e46.
- Thompson J, Higgins D, Gibson T (1994) CLUSTAL W:Improving the sensitivity of progressive multiple sequence alignment though sequence wighting, position-specific gap penalties and weitht matrix choice. Nucleic Acids Research 22(22): 4673-4680.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...