Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-6062
Mini-Review Article(ISSN: 2638-6062) 
Is PGA the Missing Piece Precision Medicine Has Been Waiting For? Volume 2 - Issue 1
Chen Yeh*
- OncoDxRx, Los Angeles, CA, USA
Received: January 23,2024; Published: January 29,2024
*Corresponding author: Chen Yeh, 150 N Santa Anita Ave., Suite 300, Los Angeles, CA 91006, USA
DOI: 10.32474/PRJFGS.2024.05.000204
Abstract
While OncoDxRx’s PGA (Patient-derived Gene expression-informed Anticancer drug efficacy) may not be as widely recognized as its counterpart next-generation sequencing (NGS), it is still considered a disruptive platform. The significance of biomarker testing in targeted therapy (or precision medicine) has been exemplified by medical guidelines. These standard of care tests provide the most valuable means of improving patient outcome and can significantly relieve long-term healthcare burden.
Due to low patient eligibility rate of targeted therapy and irrevocable drug resistance, OncoDxRx ended up developing its proprietary PGA to complement NGS and to benefit non-responder patients. PGA is a combination of liquid biopsy, in vitro gene expression profiling, and in silico computation. To improve clinical utility, the technology’s key genetic signature was derived from the patient’s own blood. In terms of operation workflow, the assay is high throughput, robust with fast turnaround. A panel of selected biomarkers is used to generate a patient’s genetic fingerprint, which then can be used to digitally screen, match and catalog potentially effective cancer drugs.
PGA features the world-first gene-to-drug revolution to help cancer patients with limited treatment options. Notably, the test’s customized report enables clinicians to have more therapy strategies in their hands than ever before.
Keywords: targeted therapy; biomarker testing; non-responders; liquid biopsy; cell-free mRNA
Introduction
Precision medicine (PM or targeted therapy) is now relevant across nearly every area of research, biomedical, pharmaceutical and otherwise. And while pathology is no exception, it may be one of the last frontiers-it’s estimated that only five to ten percent of hospitals have transitioned from glass slides to digital images. Here, we’ll explore how close we are to realizing PM-guided personalized care, along with the challenges and benefits associated with integrating new technologies into PM.
Precision medicine is central to nearly all clinical advancements
PM is critical to the development of new drugs. Historically, the field has contributed to preclinical research through target identification and delineation of drug mechanisms of action, while also assessing pharmacodynamics and toxicology. PM also connects drug discovery to translational efforts and clinical efficacy, offering insight into disease pathophysiology, patient treatment and response, and more. To illustrate, PM-associated technologies, such as biomarker testing and basket trials, are increasingly used to determine patient eligibility across diseases.
Setting the stage for precision medicine
The increased emphasis on PM and advances in technology have spurred the development of biomarker-based approaches in recent years. In the past, pathologists focused on regions of interest (ROI) from tissue samples to make a diagnosis or treatment recommendation. Now, however, there are NGS and digital PCR technologies, which can identify actionable genetic alterations from a sample to guide clinical treatment decisions. These digitized and customized reports enable direct mutation-todrug translation, leading to less inter- pathologist variability and allowing for the generation of an accessible, curated database with unlimited applications from the educational to the clinical (1, 2).
The combination of NGS and bioinformatics, which can process multiple patients with multiple molecular markers simultaneously, can be used to decipher the genotypic relationships of phenotypically distinct cell populations beyond ROIs. The vast and rich availability of information that can now be derived means there is also a need for reproducing interpretation of these complicated data sets and has led to the use of machine learning in biomarker testing.
PGA in the patient-to-drug realm
Quantitative analysis can be used to create high content gene-to-drug data through multiplexing, a proprietary liquid biopsy technology, PGA, was thus developed that shows the expression of numerous markers within the context of the complex tissue environment and at the single-patient level.
The tumor microenvironment yields endless clues about cancer, like its molecular underpinnings, relationship with the immune system, and what could potentially destroy it-but only if the vast amounts of complicated data can be accounted for and interpreted. The fusion of PGA with its in vitro assay and in silico computation is a powerful means to decode this information. These capabilities not only overcome the limitation of biomarker testing, but also decrypt correlates relevant to patient treatment especially for those non-responders. In fact, including gene expression metrics, or additional surrounding non-tumor context, in PGA also led to significantly better prediction of drug efficacy (3).
Unsupervised learning models have already been used to predict drug responses, as well as to evaluate genetic features to provide a score associated with overall survival. Still these digital approaches rely on the quality and quantity of data used to train the algorithm (4). This could be an issue as there are often differences in sample preparation, tumor profiling methods, and grading, between different cohorts and laboratories. Another obstacle is the initial capital and time investment. Although systematic computation analyses mean faster, more efficient workflows, the financial cost of equipment and software, alongside the timeconsuming nature of algorithm development, can’t be ignored.
In the era of PM, AI-assistance is already helping to match patients to the most promising treatments. In melanoma patients, adoptive T cell therapy has a greater than 50% success rate, but only in patients who can generate tumor-infiltrating lymphocytes (TIL). Feng et al utilized 7-color multispectral immunohistochemistry and unsupervised hierarchical clustering to thoroughly analyze the immune environment in melanoma tumors. The authors postulated that the immune profile could be used to select for patients with TIL capability, as well as predict who will respond to immune checkpoint inhibitors (5). Pancreatic adenocarcinoma is a cancer with a notoriously poor survival rate, typically thought to be nonresponsive to most drugs. However, the automated classification of immune cells and epithelial cells alongside single-cell level marker analysis, showed that the spatial distribution of cytotoxic T cells with relation to cancer cells is a marker of increased overall patient survival. Analyzing tumor microenvironment for T cells may help to identify which patients would most benefit from immune therapies and argue that this kind of information should be included in standard tumor pathological scoring (6).
Figure 1: A new “ build-for-purpose” test was unveiled by OncoDxRx. The innovative PGA technology aims at broadening the coverage of tailored therapies for those cancer patients currently excluded from precision medicine.
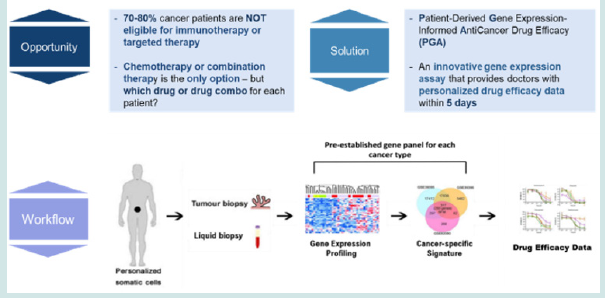
It has been well known for years that gene expression via plasma transcriptomic profiling has the potential to accelerate novel discoveries and advance our understanding of cancer biology and further precision medicine. Leveraging the exclusive liquid biopsy cell-free mRNA (cfmRNA) knowhow, PGA enables laboratories to generate actionable information from blood samples in a 5-day-turnaround workflow (Figure 1). PGA enables clinicians and clinical laboratories to access high-resolution anticancer drug efficacy results using cancer type-specific biomarkers and patient-derived gene expression signature information. PGA will take liquid biopsy cfmRNA beyond what is possible with current precision oncology testing workup and establish a new industry standard for effective and personalized treatment.
Until now, clinicians considering cancer genomic tests have had to choose between DNA mutations that provide deep coverage at the cost of resolution, or fusions/rearrangements that provide high resolution data across only a subset of targets. Not anymore. The novel and validated technique on which PGA is based allows regular laboratories to use existing qPCR instrumentation to generate unbiased, high-resolution and actionable data across individual patients who are currently out of targeted therapy or immunotherapy option, unlocking new avenues for personalized treatment.
PGA provides the ability to generate actionable report by molecularly profiling individual patients, obtaining patients’ gene expression signatures, followed by in silico screening and matching effective drugs. Oncologists can capture and recognize what the effective drugs are to better decide and optimize treatment strategy.
PGA provides a pathway for studying tumor behavior, tumor microenvironment, the immunological response, and the discovery of molecular signatures that predict drug efficacy or treatment response. It is the first in a suite of one-of-a-kind products that OncoDxRx is developing and validating which enables pointof- treatment using universally existing qPCR instrumentation.
The promise of PGA
In the clinical market space, the gene-drug relationship is believed to be the next major medical transformation. The most appealing aspects of PGA may be its capacity for benefiting patients that elude targeted therapy or immunotherapy, and its potential for discovering novel drugs, applicable not only to diagnostics, but also to the discernment of novel drug targets. PGA can also aid in stratification of patients and choosing optimal treatment regimens, particularly in complex areas such as combination therapy, where therapies continue to diversify. In this dynamic landscape, the melding of biomarker testing and PGA not only enhances our understanding of disease mechanisms but also boosts the efficiency and success rates of precision therapy, clinical trial design, and patient outcomes.
References
- Jackisch C, Hegg R, Stroyakovskiy D, Ahn JS and Melichar B et al. (2016) HannaH phase III randomised study: Association of total pathological complete response with event-free survival in HER2-positive early breast cancer treated with neoadjuvant-adjuvant trastuzumab after 2 years of treatment-free follow-up. Eur J Cancer 62: 62-75.
- Shafi S, Parwani AV (2023) Artificial intelligence in diagnostic pathology. Diagn Pathol 18(1):109.
- Yeh C (2023) A game-changer in cancer treatment. J Cancer Prev Curr Res 14(3): 65‒66.
- Baxi V, Edwards R, Montalto M, Saha S (2022) Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol 35(1): 23-32.
- Feng Z, Puri S, Moudgil T, Wood W and Hoyt CC et al. (2015) Multispectral imaging of formalin-fixed tissue predicts ability to generate tumor-infiltrating lymphocytes from melanoma. J Immunother Cancer 3: 47.
- Carstens JL, Correa de Sampaio P, Yang D, Barua S and Wang H et al. (2017) Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun 8: 15095.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...




