Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4706
Research Article(ISSN: 2637-4706) 
Potential Molecular Docking of Four Acetylcholinesterase Inhibitors Volume 2 - Issue 3
Nahd Mohamed Elmaki1, Inass A Al Sadawi2, Anton Hermann3 and Abdul M Gbaj*2
- 1Department of Pharmacy and Medical Devices, Ministry of Health, Tripoli, Libya
- 2Department of Medicinal Chemistry, University of Tripoli, Libya
- 3Department of Biosciences, University of Salzburg, Austria
Received: September 04, 2018; Published: September 10, 2018
Corresponding author: Abdul M Gbaj, Department of Medicinal Chemistry, University of Tripoli, Libya
DOI: 10.32474/DDIPIJ.2018.02.000136
Abstract
Molecular modeling attempts to study the function, structure and inhibition of the acetylcholinesterase enzyme due to the fact that the inhibition of this enzyme is importance to medical conditions such as Alzheimer’s disease, myasthenia gravis and Parkinson’s disease, and it is also important in eminent toxicological susceptibility to nerve agents. In this study we present an approach for forecasting the inhibitory activity of acetylcholinesterase (AChE) inhibitors by using docking studies. The docking studies were done on acetylcholinesterase to attain the conformation of the enzyme in water surroundings. The obtained conformation of the enzyme was used for docking with four inhibitors (physostigmine, neostigmine, pyridostigmine and rivastigmine). Docking analysis showed that hydrogen bonds and hydrophobic interactions play important tasks in the acetylcholinesterase -inhibitor complex. Subsequently, all inhibitors that bind at the catalytic site of acetylcholinesterase and their interactions with acetylcholinesterase were studied. In addition, conformation stability of acetylcholinesterase -inhibitors was studied using simulation docking technique. The complex showed that acetylcholinesterase conformation did not change significantly in the presence of the four inhibitors. This paper showed important studies on acetylcholinesterase and assists to illuminate the four inhibitors interdependencies using molecular modeling.
Keywords: Acetylcholinesterase; Molecular docking; Physostigmine; Neostigmine; Pyridostigmine; Rivastigmine
Introduction
Acetylcholine (ACh) is a neurotransmitter broadly distributed in the central and peripheral, autonomic nervous system (CNS). In the CNS, ACh performs several functions, such as memory, learning, motor control and attention. Acetylcholinesterase (AChE), one of the most essential enzymes in the family of serine hydrolases, catalyzes the hydrolysis of neurotransmitter acetylcholine, which plays an important role in memory and cognition [1-4]. It has been suggested that acetylcholinesterase (AChE) degrades the esters of choline and has a role in neurotransmission within the autonomic and somatic motor nervous systems and it is the target of action of the drugs (inhibitors) such as physostigmine, neostigmine, pyridostigmine and rivastigmine, physostigmine [1-4]. It is well known that AChE enzyme regulated the release and entrance of ACh in cholinergic fibers [5]. The function of AChE in neurological disorders like Myasthenia gravis (MG) [6], Alzheimer’s disease [7], Parkinson’s disease [8] and other ‘non-classical’ activities such as, neurite formation, network formation and cell adhesion [9] draw attention of many researches related to the medical field. The exact mechanisms of the interaction of physostigmine, neostigmine, pyridostigmine and rivastigmine with the acetylcholinesterase receptor complex still need further molecular modeling studies.
Acetylcholinesterase has extremely high catalytic activity and each molecule of AChE hydrolyze about twenty-five thousand molecules of acetylcholine (ACh) per second. The active site of AChE contains two subsites which are the anionic site and the esteratic subsite [10,11]. The mechanism of action and structure of AChE have been obtained from the crystal structure of the enzyme which were studies by different research groups [12- 14]. The anionic subsite housing the positive quaternary amine of the acetylcholine in addition to other cationic inhibitors and substrates. The substrates which have cationic charge do not bound by the negatively charged amino acid in the anionic site, but the binging is performed through interaction of fourteen aromatic residues that available close to the active site [15-17]. The active site of AChE is located four angstroms from the substructure of the molecule [17]. The acetylcholine is hydrolyzed in the esteratic subsite and produces acetate and choline. The active site contains mainly a catalytic triad of three amino acids which are: serine 203, histidine 447 and glutamate 337 [18]. The hydrolysis reaction by AChE on carboxyl ester produces acyl-enzyme and free choline. Subsequently, the acyl-enzyme undertakes nucleophilic attack by a water molecule, supported by the histidine 447 moiety and the releasing acetic acid and regenerating the free enzyme again [18]. The main aims of the paper are: virtual screening approach, pharmacophore modeling, molecular docking, and consensus binding affinities of acetylcholine and four well know inhibitors. The obtained results will be used as a guide to identify and design novel AChE inhibitors with higher selectivity.
Materials and Methods
Molecular docking
The starting geometry of physostigmine, neostigmine, pyridostigmine and rivastigmine was constructed using chem3D Ultra (version 8.0, Cambridgesoft Com., USA). The optimized geometry of the four inhibitors with the lowest energy was used in the molecular docking. The crystal structures of human acetylcholinesterase in a complex with a transition-state analogue were downloaded from the Protein Data Bank (https://www. rcsb.org/structure/5hfa). The molecular dockings of the four inhibitors with human acetylcholinesterase was accomplished by AutoDock 4.2 software from the Scripps Research Institute (TSRI) (http://autodock.scripps.edu/). Firstly, the polar hydrogen atoms were added into human acetylcholinesterase and the four inhibitors molecules. Then, the partial atomic charges of the human acetylcholinesterase and the four inhibitors molecule were calculated using Kollman methods [19]. In the process of molecular docking, the grid maps of dimensions (62Å X 62Å X 62Å) with a grid-point spacing of 0.376Å and the grid boxes is centered. The number of genetic algorithms runs, and the number of evaluations was set to 100. All other parameters were default settings. Cluster analysis was performed on the results of docking by using a root mean square (RMS) tolerance of 2.0Å, and this was dependent on the binding free energy. Lastly, the dominating configuration of the binding complex of the four inhibitors and human acetylcholinesterase with minimum energy of binding can be determined.
Results and Discussion
Molecular Docking Analysis
The modeling study was performed in this paper showed great interactions between physostigmine, neostigmine, pyridostigmine and rivastigmine and human acetylcholinesterase. The binding energies of the four inhibitors and human acetylcholinesterase were shown in Table1. The geometry of docking obtained with acetylcholine and pyridostigmine with human acetylcholinesterase as shown in (Figure 1A & B), respectively. The four inhibitors were able to form hydrogen bonds (HBs) with the amino acid residues of the enzyme, pi-pi stacking and Pi-alkyl interaction (Table 1). In addition, the molecular docking results showed that other amino acids residues are involved in the interactions with the four inhibitors. The binding energies scores as shown in Table 1 were calculated by adding up a set of weighted empirical energies including van der Waals forces (VDW), electrostatic, hydrogenbonding, desolvation, entropy, and hydrophobicity. A good number of pi‒pi interactions were observed (Table 1) that show the bond distance varying between 3.40 Å to as far as 3.95 Å which is consistent with the results obtained by Avasthi et al. whom indicated that the ideal bond distance in pi‒pi interactions are within the range 3.30–4.00 Å [20]. In addition, in order to provide another clear reason of the activity difference in terms of dynamic behavior, distances Pi-sigma monitor interactions between Ser203 and ligands were determined and compared during the simulation time (Table 1). The distances obtained were similar to the literature where the distance of Pi-sigma interaction less than 0.5nm [21]. This indicated that the π-sigma interaction could also be one of the key interactions to elucidate the activity dissimilarity in terms of dynamic behavior. The docking results obtained in Table 1 with the acetylcholine and the various inhibitors provided insights into the essential structural elements and motifs central of the catalytic mechanism of the acetylcholinesterase. It has been reported that the X-ray analysis of the structural features of AChE has a narrow, long, hydrophobic gorge which is about 20 Å deep [11,12]. The AChE has a catalytic triad consisting of His447, Ser203, and Glu334 [22] situated in the active site of the narrow deep gorge, the lining of which contains mostly aromatic residues that form a narrow access to the catalytic Ser203 [22]. In addition to that a peripheral anionic site containing an additional set of aromatic residues Trp286, Tyr124, Tyr72, Tyr341, and Asp74 [22] was reported which is located at the border of the gorge and affords a binding site for allosteric inhibitors and modulators. The binding between the acetylcholine and the four inhibitors with the enzyme is characterized by cation-π interactions between the protonated nitrogen’s and the preserved aromatic residues, phenylalanine and tryptophan. Furthermore, π-π stacking between the aromatic moieties of the inhibitors and the aromatic amino acids as shown in Table 1 has critical roles in binding and this consistent with the literature as described by Shin Hua et al [18]. With the current obtained data, computational methods have become essential to biological investigations. Here we have used computational approach to further understand the mechanism of interactions and binding affinity between AChE with drug molecules. The present analysis permits us to draw the number of conclusions. The computational methods on the catalytic site help the researcher to design new drug molecules. The molecular docking programs are helpful in understanding the interaction between the AChE with diverse lead/drug molecules. Our analysis also demonstrates that the four inhibitors could be the prospective lead molecule for the inhibition of AChE. Hence docking results of acetylcholine and the four inhibitors could be used as the template for designing therapeutic lead molecule. We hope that the originality and success of the computational efforts in this paper offer fruitful promise for the future predictions of finding new inhibitors which could results into enormous reductions in therapeutics development time.
Table 1: Binding and intermolecular energies from molecular docking for compounds acetylcholine, physostigmine, neostigmine, pyridostigmine and rivastigmine.
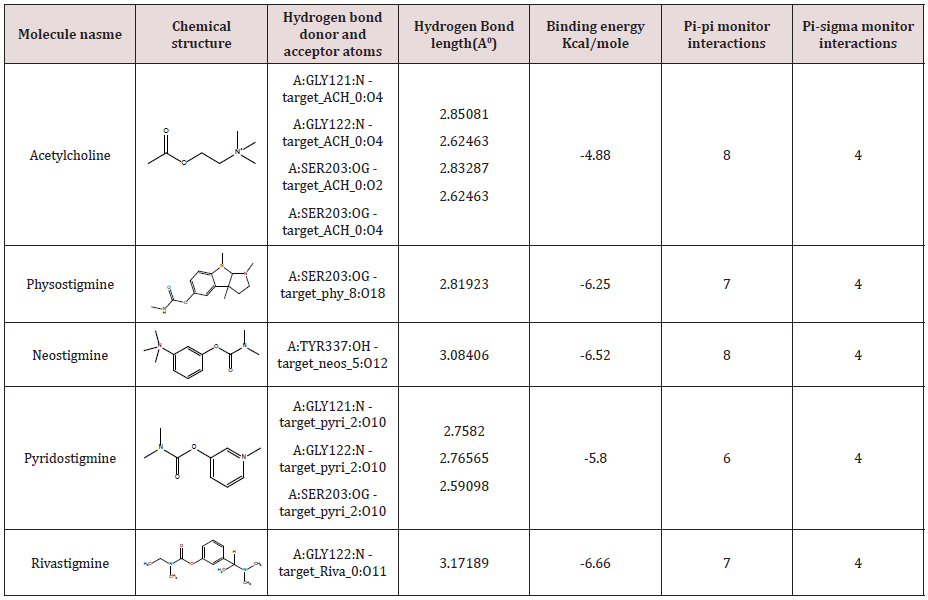
Figure 1: Three-dimensional representation of acetylcholine (A) and pyridostigmine (B) interacting with active site of target macromolecule human acetylcholinesterase.
Conclusion
ILs are receiving more and more attention every day both in academic research and commercial applications and they seem as good replacements for volatile organic solvents. However, there is a discussion about the greenness of the ILs due to their incomplete physical, chemical and toxicological data. most of the researchers will continue to work with this new solvent, the developments of new applications utilizing ILs will increase rapidly and the number of publications will rise exponentially in the future. The present costs of the ILs are quite prohibitive in many probable commercial applications. However, there are hopes that in the near future, the cost/benefit figures of the ILs will bring economic viability to their more common use. The development of pharmaceutical applications using IL-based methodologies requires a deep understanding of ILs both in terms of their macroscopic properties and also at the molecular level, because structural aspects have been shown to play a crucial and unexpected role in a large number of situations. Despite the fact that ILs can no longer be considered a new field, the large diversity of combinations of cations and anions producing novel ILs with new specific properties is astonishing.
References
- Augustinsson KB, Nachmansohn D (1949) Distinction between Acetylcholine-Esterase and Other Choline Ester-splitting Enzymes. Science 110(2847): 98-99.
- Yu QS, Holloway HW, Luo W, Lahiri DK, Brossi A, et al. (2010) Long-acting anticholinesterases for myasthenia gravis: synthesis and activities of quaternary phenylcarbamates of neostigmine, pyridostigmine and physostigmine. Bioorg Med Chem 18(13): 4687-4693.
- Beri V, Gupta R (2007) Acetylcholinesterase inhibitors neostigmine and physostigmine inhibit induction of alpha-amylase activity during seed germination in barley, Hordeum vulgare var. Jyoti. Life Sci 80(24-25): 2386-2388.
- Jonecko A (1963) [On the Inhibition of the Histochemical Acetylcholinesterase Reaction on Motor End-Plates by Injections of Neostigmine and Physostigmine]. Acta Histochem 16:375-381.
- Kitz RJ, Braswell LM, Ginsburg S (1970) On the question: is acetylcholinesterase an allosteric protein? Mol Pharmacol 6(2): 108- 121.
- Seifert SA, Eldefrawi ME (1974) Affinity of myasthenia drugs to acetylcholinesterase and acetylcholine receptor. Biochem Med 10(3): 258-265.
- Agatonovic-Kustrin S, Kettle C, Morton DW (2018) A molecular approach in drug development for Alzheimer’s disease. Biomed Pharmacother 106: 553-565.
- Sawada H, Oeda T, Kohsaka M, Umemura A, Tomita S, et al. (2018) Early use of donepezil against psychosis and cognitive decline in Parkinson’s disease: a randomized controlled trial for 2 years. J Neurol Neurosurg Psychiatry.
- Paraoanu LE, Layer PG (2008) Acetylcholinesterase in cell adhesion, neurite growth and network formation. FEBS J 275(4): 618-624.
- Hasin Y, Avidan N, Bercovich D, Korczyn A, Silman I, et al. (2004) A paradigm for single nucleotide polymorphism analysis: the case of the acetylcholinesterase gene. Hum Mutat 24(5): 408-416.
- Botti SA, Felder CE, Lifson S, Sussman JL, Silman I (1999) A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J 77(5): 2430-2450.
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, et al. (1991) Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science 253(5022): 872-879.
- Silman I, Harel M, Axelsen P, Raves M, Sussman JL (1994) Threedimensional structures of acetylcholinesterase and of its complexes with anticholinesterase agents. Biochem Soc Trans 22(3): 745-749.
- Sussman JL, Harel M, Silman I (1993) Three-dimensional structure of acetylcholinesterase and of its complexes with anticholinesterase drugs. Chem Biol Interact 87(1-3): 187-197.
- Radic Z, Gibney G, Kawamoto S, MacPhee-Quigley K, Bongiorno C, et al. (1992) Expression of recombinant acetylcholinesterase in a baculovirus system: kinetic properties of glutamate 199 mutants. Biochemistry 31(40): 9760-9767.
- Ordentlich A, Barak D, Kronman C, Ariel N, Segall Y, et al. (1995) Contribution of aromatic moieties of tyrosine 133 and of the anionic subsite tryptophan 86 to catalytic efficiency and allosteric modulation of acetylcholinesterase. J Biol Chem 270(5): 2082-2091.
- Ariel N, Ordentlich A, Barak D, Bino T, Velan B, et al. (1998) The ‘aromatic patch’ of three proximal residues in the human acetylcholinesterase active centre allows for versatile interaction modes with inhibitors. Biochem J 335 (Pt 1): 95-102.
- Lu SH, Wu JW, Liu HL, Zhao JH, Liu KT, et al. (2011) The discovery of potential acetylcholinesterase inhibitors: a combination of pharmacophore modeling, virtual screening, and molecular docking studies. J Biomed Sci 18: 8.
- Tiwari R, Mahasenan K, Pavlovicz R, Li C, Tjarks W (2009) Carborane clusters in computational drug design: a comparative docking evaluation using AutoDock, FlexX, Glide, and Surflex. J Chem Inf Model 49(6): 1581- 1589.
- Avasthi K, Shukla L, Kant R, Ravikumar K (2014) Folded conformations due to arene interactions in dissymmetric and symmetric butylidenelinker models based on pyrazolo[3,4-d]pyrimidine, purine and 7-deazapurine. Acta Crystallogr C Struct Chem 70(Pt 6): 555-561.
- McGaughey GB, Gagne M, Rappe AK (1998) pi-Stacking interactions. Alive and well in proteins. J Biol Chem 273(25): 15458-15463.
- Ordentlich A, Barak D, Kronman C, Flashner Y, Leitner M, et al. (1993) Dissection of the human acetylcholinesterase active center determinants of substrate specificity. Identification of residues constituting the anionic site, the hydrophobic site, and the acyl pocket. J Biol Chem 268(23): 17083-17095.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...