Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4706
Research article(ISSN: 2637-4706) 
Designing Effective Drugs against Leukemia: Current Obstacles, Lead Discoveries and Drug Development Strategies Volume 4 - Issue 2
Wanpeng Sun*
- Faculty of Science, University of Alberta, Canada
Received: May 14, 2024; Published:May 22, 2024
Corresponding author:Wanpeng Sun, Independent Researcher, Newfoundland, Canada
DOI: 10.32474/DDIPIJ.2024.04.000185
Abstract
Leukemia is a serious cancer threatening all ages of people globally. This article reviewed the advantages and disadvantages of current therapies against Leukemia. Moreover, we identified a lead compound. Through in-depth analysis, we revealed 13 novel molecules, who could be promising drug candidates for future structure-based drug development. Further, we proposed a representative workflow, named Sun’s Flow Chart in Drug Discovery, to standardize the drug development process. While “all roads lead to Rome”, the Sun’s Flow Chart is one of most simple, rational and straightforward approaches. In addition, standardizing a process could provide help hints for the emerging human-AI complex and automation applications.
Keywords: Leukemia; Rational Drug Design; Sun’s Flow Chart in Drug Development; Sun’s Paradox in Drug Discovery; Drug Development; Structure-based Drug Design (SBDD); Lead Compound; Drug Discovery; Topoisomerase; Cancer Therapeutics
Introduction
Leukemia is a serious cancer threatening all ages of people world widely. In North America, there are more than 200,000 patients suffering from leukemia. Annually, around 34,000 and 4,000 new cases for leukemia happen in USA and Canada respectively. In developed countries, around 1 % populations die from leukemia.
Leukemia is the cancer that starts in blood-forming tissue such as the bone marrow and causes large numbers of blood cells to be produced and enter the bloodstream. It is characterized by a dramatic increase in white blood cells. This leads to a reduced number of red blood cells and planets. The lack of them leads to deficiency of red blood cells and excessive bleeding. The deficiency of red blood cells triggers a series of severe pathological states, such as hypoxia and dyspnea. Moreover, the abnormally proliferated white blood cells lose their normal functions resulting in frequent infections. Furthermore, a series of symptoms are associated with leukemia. For example, leukemia cells can accumulate in almost any part of the body, leading to unusual swelling and pains. It is even worse if they accumulate in the brain. In this case, patients can suffer headaches, confusion, and seizures.
Leukemia can be divided into the acute type and the chronic type, the latter of which will convert to the acute type eventually. If untreated, patient will die rapidly in the acute type. Leukemia can influence either myeloid stem cells or lymphoid stem cells. In the former case it is called myelogeneous leukemia while in the latter case it is called lymphogeneous leukemia. There are both acute and chronic types in either myelogeneous leukemia or lymphogeneous leukemia. The causes for leukemia are diverse, such as radiations, chemicals, virus, and genetical factors. There are also many cases in which no known factors can be identified as the cause. To date, no specific cause-type relationships have been discovered yet.
Challenges and Obstacles
There are many types of therapies in the treatment of leukemia. The most common and effective treatment is chemotherapy, in which anti-cancer drugs are applied to remove leukemia cells. Most drugs in the chemotherapy target at nucleotides, since leukemia cells are usually fast dividing and are very sensitive to DNA damage. Doxorubicin, which intercalates into the space between adjacent DNA base pairs and disrupts DNA functions in leukemia cells, is the most effective drug for leukemia treatment (e.g. Taylor et al., 1982; Bern et al., 1987; Pan et al., 2002) [1-3]. In contrast, another type of nucleotide-targeting drug, alkylating agents, which form covalent bonds in nucleotides and even cross-link two nucleotides, are not used to treat leukemia because they actually induce leukemia (Reimer at al, 1977),[4] although they are effective in many cancers like ovarian cancer. There are also a few drugs which target proteins. Vincristine targets microtubule polymerization to affect cell division (Kempin et al., 1982; Jordan et al., 1985) [5,6]. Etoposide (VP-16®) and teniposide (Vumon®), may inhibit topoisomerases and are used to treat leukemia; however because they also bind to the DNA part of the topoisomerase-DNA complex, DNA could also be their target. Further, they are found to induce secondary leukemia (Relling et al., 1998) [7].
New drugs are being developed to target the key proteins in the signal pathways of leukemia cells, such as imatinib targeting at tyrosine kinase (Deininger et al., 2005) [8] In chemotherapy, drug resistance (even cross-resistance) happens quite often, and it is always stated that “the effect of anti-cancer therapy is limited by the development of drug resistance” (Blagosklonny et al., 1999) [9]. In this situation, a second set of drug(s) usually have to be administrated. Radiotherapy is often used in combination with chemotherapy. Biotherapy is also used to inhibit leukemia cell proliferation by interferon. Surgery is sometimes applied to remove abnormally enlarged spleen to help increase the amount of red blood cells and avoid compressing other organs. All those kinds of therapy can damage normal cells, and cause notorious side effects, such as hair loss, gastrointestinal diseases, anemia, leucopenia and thrombopenia. Bone marrow transplantation is sometimes applied to stimulate new bone marrow growth when chemotherapy strongly damages the bone marrows and blood cells. However, unless the bone marrow is obtained from the patient himself/ herself, immunological responses are serious concerns.
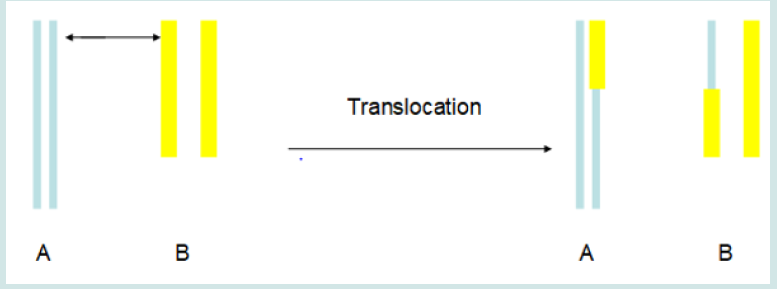
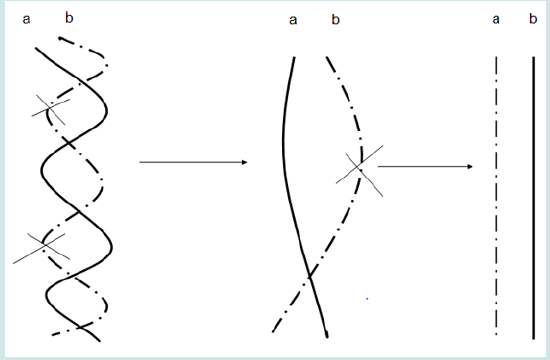
It is worthy of noting that many of reports have revealed that leukemia is associate with translocation in many cases (e.g. Nucifora et al., 1993; Licht et al., 1995; Huntly et al., 2001) [10-12]. During translocation, two portions in two chromosomes exchange positions with each other (Figure 1). Translocation activates an oncoprotein and leads to lykemogenesis (Mistry et al., 2005) [13]. It is revealed that topoisomerase II mediates the process of translocation (Felix et al., 1995; Kanoe et al., 1999) [14,15]. Some inhibitors of topoisomerase II, such as VP-16® and Vumon®, are effective antileukemia drugs. Topoisomerases are enzymes that change the topology of DNA. They regulate the three-dimensional structure of DNA and alter supercoiling chromosomal DNA to allow replication and transcription (Wang, 1996) [16]. There are at least four types of topoisomerases. Topoisomerase I can cut one strand of doublehelix DNA; topoisomerase II can cut both strands; topoisomerase III mediates recombination; topoisomerase IV meditates the sorting of newly-replicated chromosome. Some inhibitors of topoisomerase I, such as topotecan and irinotecan, are also being recognized as anticancer drugs to inhibit DNA replications in cancer cells. (Figure 1).
Inhibitors of topoisomerase II are being tested for antileukemia treatment. These inhibitors are derivatives of antibiotic drugs, while no endogenous topoisomerase II inhibitor have been defined yet. Derived from podophyllotoxin found in Mayapple, VP-16® and Vumon®, are approved to treat leukemia clinically. However these two drugs also cause drug-resistance, which renders long term treatment by these drugs inefficient. It is worthy of noting that not all inhibitors of topoisomeraes have to specifically target at topoisomerases. For example, doxorubicin inhibits the activity of topoisomerase II by intercalating into DNA (Schneider et al., 1979) [17]. There are different mechanisms underlying the inhibition of topoisomerase II. Topoisomerase II works to unwind the supercoiled DNA (Figure 2). To unwind, topoisomerase II binds to DNA to form the cleavable DNA-topoisomerase II complex, utilize the energy of ATP hydrolysis, cut a segment in one DNA molecule, allow the segment in another DNA molecule to pass through, and religate the cut segment. Topoisomerase II could be inhibited by interrupting any steps of its action (Larsen et al., 2003) [18]. Some inhibitors stabilize the DNA-topoisomerase II complex to delay the action of this enzyme. These kinds of inhibitors are traditionally called topoisomerase poisons, such as doxorubicin. Other inhibitors act on other catalytic steps, and those inhibitors are called catalytic inhibitors, such as novobiocin and merbarone. (Figure 2).
Figure 2: Schematic chart shows the unwinding process of two DNA molecules (a and b) in one chromosome by topoisomerase II. X represents the bond-cutting actions of topoisomerase II.
Novobiocin inhibits topoisomerase II by interrupting ATP binding (Gormley et al., 1996) [19]. It enhances the anti-tumor effects of alkylating agents by inhibiting topoisomerase II action on repairing cross-linking of DNA (Eder et al., 1987) [20], and enhances the anti-tumor effects of etoposide and teniposide by accumulating them inside cells (Rappa et al., 1993) [21]. However, novobiocin itself serves as an antibiotic rather than an anti-cancer agent. It has multiple targets in cells, such as RNA polymerases (Gottesfeld, 1986). [22] Therefore, it is not a good candidate to develop for leukemia therapy.
Another exogenous topoisomerase II inhibitor, merbarone, specifically inhibits topoisomerase II (Figure 3); Drake et al., 1989) [23]. It reduces the topoisomerase II activity on cutting DNA (Fortune and Osheroff, 1998) [24], and consequently delays the movement of the replication fork in S phase of mitosis (Wang et al., 2007) [25]. Merbarone has been demonstrated to be an effective anti-leukemia agent in animal model (Glover et al., 1987) [26]. However, in clinical trials, it fails to display significant anti-cancer activity (Kraut et al., 1992; Malik et al., 1997) [27,28]. (Figure 3).
Drug Discovery
Lead Compound
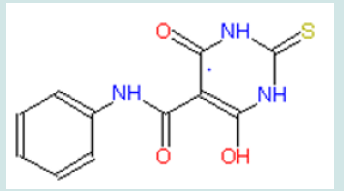
Recently quinolone derivatives specifically inhibiting topoisomerase II have been identified (Figure 4). The inhibition happens in both purified enzyme and in living cells (Wentland et al., 1993, Permana et al., 1994; Coughlin et al., 1995; Tabarrini et al., 1999) [29-32]. In these studies, quinolone derivatives have been demonstrated to have anti-cancer actions. Unlike doxorubicin that binds to DNA, quinolone derivatives do not directly bind to DNA (Coughlin et al., 1995) [31]. Among those quinolone derivatives, WIN 58161, (S)-10-(2,6-Dimethyl-4-pyridinyl)-9-fluoro-3-methyl- 7-oxo-2,3-dihydro-7~-pyrido[l,2,3-de]-[l,4]benzothiazine-6- carboxylic acid, attracts my attention. No drug resistance to WIN 58161 has been found in leukemia cells (Coughlin et al., 1995). It displays significant anti-cancer activities (Reuman et al., 1991; Coughlin et al., 1995) [33,31]. In the concentration of 23 μM, it kills 50% leukemia cells in one hour, while same concentrate of another drug shows no obvious effects on the parallel group. In vitro, it effectively induces the death of P388 leukemia cells. It also shows anti-leukemia activities in the animal model of mice. This anti-leukemia activity is specifically mediated by topoisomerase II. The specifically targeting on the protein of topoisomerase II is beneficial to further studying drug-receptor interaction by threedimensional modeling. Therefore, we will use WIN 58161 as the lead compound. (Figure 4).
Figure 4: Structures of representatives of four quinolone derivatives, all of which show anti-leukemia activities.
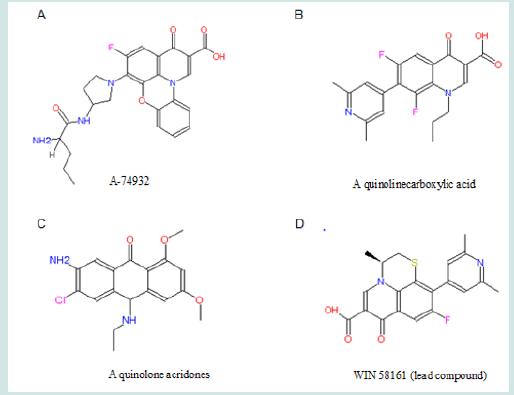
The combined structure of a benzene ring and a pyridine ring is always associated with the anti-leukemia activities of quinolone derivatives (Chu et al., 1992; Wentland et al., 1993; Coughlin et al., 1995; Tabarrini et al., 1999 [34,29,31,32]). Further, the extended 10-pyridinyl group in WIN 58161 could benefit the inhibitory action on topoisomerase II (Coughlin et al., 1995) [31]. Interestingly, Wetland et al. (1993) [29] also revealed an anti-leukemia topoisomerase II inhibitor (a quinoline carboxylic acid) which has the benzene-pyridine combined structure and the extended pyridinyl group (Figure 4B). However, when both comparing to the anti-cancer effect of V-16, WIN 58161 is much more potent than the quinoline carboxylic acid. Additional linking between the benzene ring and the pyridinyl ring by a third ring structure might play a role to enhance the inhibition of topoisomerase II. This tricyclic structure makes quinolone derivatives more effective on inhibiting type II topoisomerase (Lesher, 1989) [35]. The thiazine structure in WIN 58161 is an optimized configuration for the third ring structure. Substituting the sulfate atom by oxygen dramatically reduces the inhibitory effect (Coughlin et al., 1995) [31]. The stereoisomer of WIN 58161, which have the 3-methyl pointing into the opposite direction of thiazine plane, shows little, if any, activity to inhibit topoisomerase II and leukemia (Coughlin et al., 1995) [31].. Overall, given its target selectivity, anti-leukemia activity, lack of drug resistance, and specific structures which inhibits topoisomerase II, WIN 58161 is selected as the lead compound in this project.
Drug Development Strategy
Structural modifications will be made based on the current structure of the lead compound, WIN 58161. These modifications intend to improve the receptor binding, and acquire more information about how WIN 58161 binds to topoisomerase II. Some substitutions will also be made in order to identify the functional groups.
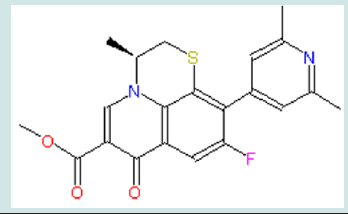
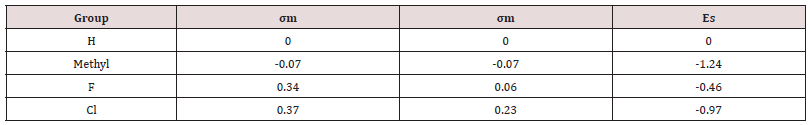
Present in many anti-leukemia compounds such as A-74932, the carboxyl group in quinolone seems necessary for receptor binding. To further confirm this idea, a compound (compound 1) with an ester modification on the carboxyl group will be tested (Figure 5). This modification is expected to decrease the receptor binding capability and demonstrate that the carboxyl group is essential. If the result turns out as expected, electron withdrawing groups (EWGs) can be added to the pyridinyl ring to examine their interaction with the carboxylic group (Figure 6). EWG groups tend to increase Ka and therefore decrease pKa (-logKa) and increase acidity of carboxylic acid by stabilizing its negative state. Electronic withdraw effects will be expected to make carboxylic group more electronegative and therefore better for binding. The inductive effects can be made either through making electron density in the pyridinyl ring spread toward EWG to affect attached carboxylic group or through directly withdrawing electrons from the nearby carboxylic group toward the halogen atom. At the present point, it is hard to predict which substituent, F or Cl, has stronger inductive effect to benefit binding since Hammett constant is not available for ortho-subsitution due to steric influence.
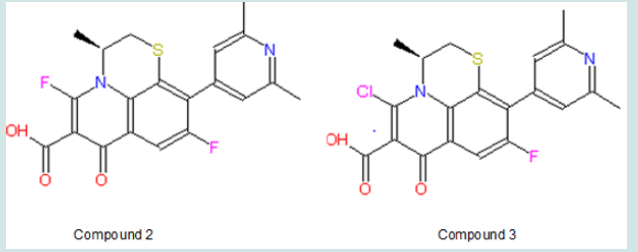
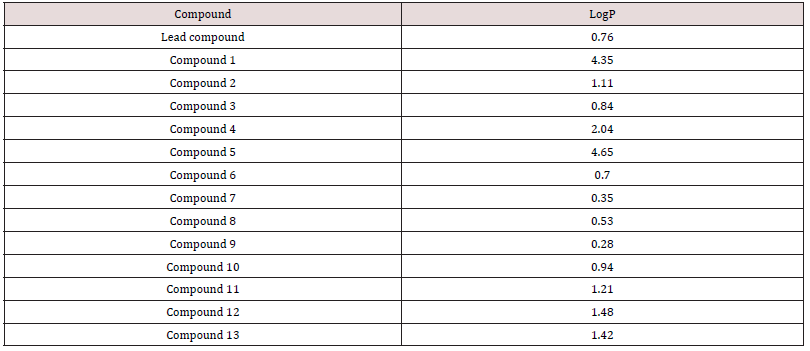
The σm is 0.37 and 0.34 for Cl and F respectively, and σp is 0.23 and 0.06 for Cl and F respectively. In terms of σm and σp, Cl seems to have more influence to spread electrons in aromatic rings. Nevertheless, F has relatively higher electronegativity (3.98) than Cl (3.16), thus F might have more electronic effect on nearby carboxylic group. Also, the steric effect may be considered since halogen and carboxylic group are close to each other. Taft’s steric factor (Es) for F and Cl is -0.46 and -0.97 respectively (Table 1), referring to H (Es for H is 0). Therefore, chloride may have stronger steric influence than fluoride. Besides, F or Cl atom might form hydrogen bond with the receptor to help binding. On the other hand, in case that there is size limitation on the binding pocket, adding halogen atom, especially chloride with larger size, may make the compound hard to fit into the pocket so as to reduce binding capability. Additionally, this modification will bring the predicted logP value from 0.76 of the lead compounds to 1.11 and 0.84 of compound 2 and 3 respectively (Table 2); LogP is predicted on-line by Interactive Analysis; same as below predictions). In compound 2 and 3, intra-molecular hydrogen bond could be formed between fluoride/chloride and hydrogen in the carboxylic group to make them less hydrophilic. As a result, their increased lipid solubility helps them pass cellular membranes, as well as blood-brain barrier in case the leukemia spreads into brain. (Table 1), (Table 2), (Figure 5), (Figure 6).
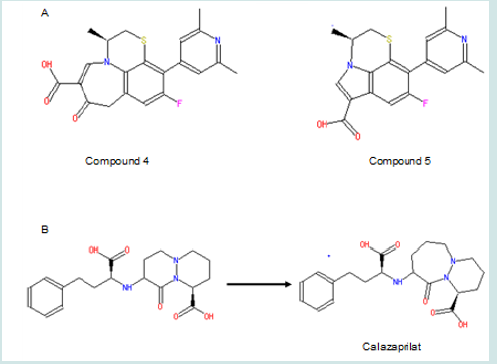
Moreover, compound 4 and 5 with the modifications on the size of the pyridinyl ring in quinolone will be tested (Figure 7A). This aims to find if the carboxylic group will be placed into the optimal orientation for receptor binding. Changing ring size is a good way to optimize the position of its substituent(s) for binding. For example, cilazaprilat has been successfully developed through ring expansion strategy (Figure 7B). In that case, increasing the size of ring 1 from six membered to seven membered put the carboxylic groups to optimal orientation for receptor binding. No matter whether the ester compound 1 can change the binding activity as expected, these modifications are worthy of being tested since they also help to find whether the six-member structure in this pyridinyl ring is essential for binding. Moreover, the logP values are predicted to increase to 2.04 and 4.65 in compound 4 and 5 respectively. The increased lipid solubility is helpful for these agents to travel through not only the cell membrane but also the nuclear membrane, since topoisomerase II is located inside nuclei. It is noteworthy that, in compound 4, because intra-molecular hydrogen bond can form between the hydrogen in the carboxylic group and the oxygen linking to the pyridinyl ring, the logP value could be underestimated by the current method. This kind of underestimation also happens to some aromatic derivatives such as the one shown in (Figure 8); in this compound, the experimental logP value is 1.27, while the predicted value is only -1.30. Thus, compound 4 may be more lipophilic to pass double membranes than predicted. Lipophility also delays the drug metabolism which largely depends on hydrolysis, makes the drug stay longer inside body with longer action time. Alternatively, the ester compound 1 might turn out to enhance the binding capability; this means the ester group also benefits binding. Then compound 4 and 5 are still worthy of being tested to find if they can place the ester group into the optimal configuration for receptor binding. (Figure 7), (Figure 8)
Figure 8: Structure of an aromatic compound whose logP value is underestimated by the current predicting method.
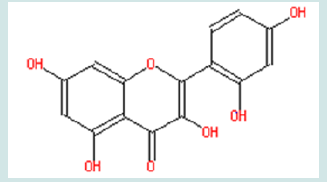
On the other hand, even if it turns out that compound 1 does not affect the receptor binding at all, synthesized compound 1 is not a waste. Its ester modification dramatically increases the logP five-fold (logP = 4.35). This increases lipid solubility and makes it easier to pass through cellular membranes. Compound 1 can still be tested in future bioassay system, in which compound 1 will be converted back to the lead compound by the intracellular esterase, to compare and evaluate activities with other potential drugs. Moreover, compound 1 might delay metabolizing process. Although quinolone are hardly metabolized inside body and are mainly eliminated by kidney in urine excretion (Vree et al., 1986) [36], there are still some predictions which can be made for the metabolism of WIN 58161. In phase I, it is likely metabolized by cytochrome P450 through methyl hydroxylation, and then in phase II, it is likely metabolized by glucuronic acid conjugation and/or sulfonation in the hydroxyl group or amino acid conjugation in the carboxylic group. Compound 1 changed carboxylic group to ester and avoid the possibility of amino acid conjugation.
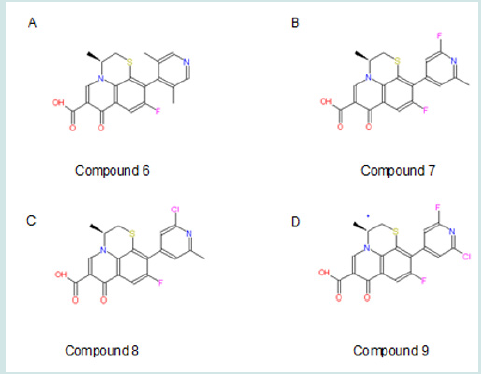
The extended 10-pyridiny ring in the lead compound could also be modified. Compound 6 can be made to discover the conformational preference of this ring (Figure 9; A). Theoretically, in the original lead compound, the extended meta-dimethyl pyridinyl ring and the benzene ring are expected to be in the same plane. After moving the methyl groups closer to the benzene ring, the steric bulk will be adequate to make the 10-pyridinyl ring outof- plane. This ortho-dimethyl pyridinyl ring is expected to maintain to a 90° angle with the benzene ring. This will help determine an optimal steric conformation to best fit the receptor. Some drugs, such as clonidine, are made more effective (Stahle, 2000) [37]by adjusting rings to non-planar. Alternatively, the pyridinyl ring can be modified by substituting methyl groups with isostere groups such as chloride and fluoride groups (Figure 9; B, C, D). Compounds 7-9 are made by this kind of substitution with halogen EWG(s). The modifications help us to understand whether methyl or dimethyl group(s) are essential for receptor binding. If the methyl group is necessary for binding, compound 7 ,8 and 9 will dramatically decrease binding capability; if only compound 9 decreases binding capability, it means only one methyl are necessary for binding. If methyl group is not necessary for binding, the pyridinyl ring itself is important for binding. Pyridine is a base with electronegative nitrogen atom that tends to form hydrogen bind with another molecule (Krygowski et al., 2005) [38]. EWG subsituent(s) may increase receptor binding. The halogen subsituent has higher σ values than the original methyl group, whose σm and σp are both -0.07. The halogen affects binding by withdrawing electrons in the aromatic pyridinyl ring. This will reduce the pKa and make N atom more electronic negative with more tendency to form hydrogen bond with the receptor. Further, halogen substitution can increase the duration of drug activity by metabolically blocking the methyl hydroxylation effect of cytochrome P450 in phase I metabolism and subsequently delaying the phase II metabolism. Besides, weak hydrogen bonds might form between halogen atom and the binding pocket. In addition, the Es values is different even in these isostere substitutions (Table 1). After receptor docking is examined in the second part of this project, the relationship between Es and binding will be clarified. Nevertheless, halogen atom has potential toxic effects as showed in many insecticides like DDT, which can also inhibit leukemia (Silinskas and Okey, 1975) [39]. Therefore, carefully screening is required in future bioassay system to find the appropriate compound with minimal adverse side effects. In addition, all these modifications in the extended 10-pyridinyl ring slightly reduce the predicted logP values to 0.70, 0.35, 0.53, and 0.28 in compound 6, 7, 8, and 9 respectively. This may cause better absorptions of these agents in the gastrointestinal tract (GIT) and increases the bioavailability of oral intake, whereas this slightly decreases the membrane penetration. (Figure 9).
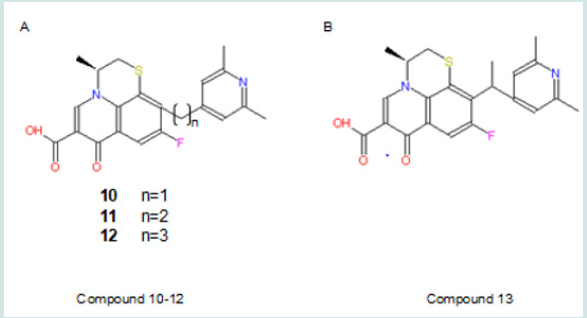
Alkyl chain extension is an effective way to develop new drugs such as the most popular ACE inhibitor, Enalaprilat (Lin and Lu, 1997) [40]. In this study, chain extension could be made between the 10-pyrindine ring and the benzene ring. Compound 10-12 can be developed as showed in Figure 10 A. These modifications intend to explore the size of the binding pocket, explore extra binding regions, and find best-fit size of the alkyl chain. The expected trends are 1) that the binding capability will increase when the size of the alkyl chain increases, if there are extra binding sites and extra space in binding pocket; 2) that binding capability will increase and decrease with the increasing size of the chain if there are available extra binding sites and extra limited room; 3) that binding capability will decrease with increasing size of the chain, if the binding pocket has already been fit tightly by the lead compound. Also, a branch can be made on the extended chain as compound 13 showed in Figure 10 B. This will examine if there are other steric limitations for the alkyl chain beside its length. In addition, this modifications increase logP values (0.94, 1.21, 1.48 and 1.42 in compound 10, 11, 12, and 13 respectively). This will improve the lipid solubility and help them pass across two membranes of cell and nuclei. (Figure 10).
Overall, drug candidates of forming ester, adjusting ring size, making substitution, shifting group positions, and chain extension/ branching, are designed. For further research, binding activity will be examined. The relationship between binding capability and σ, Es, and P will be evaluated to facilitate further drug design. These will provide valuable information about receptor binding, and help to design the best-fit candidate for receptor binding, and benefit the development of anti-leukemia therapy.
Workflow in Drug Development
Furthermore, we propose a representative workflow, named Sun’s Flow Chart in Drug Discovery, to standardize drug development process. While “all roads lead to Rome”, the Sun’s Flow Chart is one of most simple, rational and straightforward. In addition, standardizing a process could provide help hints for the emerging AI and automation applications.
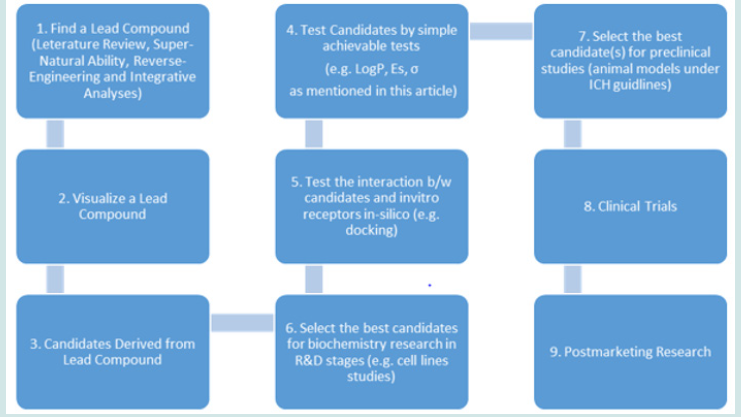
Historically, among every 5000 compounds being studies in labs, only one got approval from FDA for clinical trials. During clinical trials, more candidates failed. Thus, the cost of developing a modern drug is huge: generally, a successful drug costs 1 billion dollars from research to approved marketing (after clinical trial stages). For difficult diseases like cancer, the costs are often doubled or even tripled. In Sun’s Flow Chart (Figure 11), the first step is to identify an ideal lead compound. This step is crucial to reduce the huge cost in the downstream drug discovery. This step is usually achieved by analyzing the results of previous experiments/data, e.g. literature reviews. For example, in this article, we use literature review to identify a promising lead. In addition to this approach, it can also be achieved by reverse engineering and supernatural prediction. Due to intellectual property (IP) issues, reverse engineering approach is not recommended. In a previous publication (Sun, 2024) [41], we proposed the Sun’s Paradox: in ancient time, there are no modern knowledge and equipment, how can human beings randomly screen millions of candidates to find medicine? This could be achieved by supernatural power processed by humans, just like the case of Kekule who discovered the benzene ring structure in dream. Despite of many ways of finding a lead compound, serious integrative analyses are always required to uncover the ideal lead. The second steps are visualizing a lead compound once it has been identified. It is like an interview. This step is not only critical for a medicinal chemist to design the derived compounds, but also give scientists a moment to feel a candidate by heart (intuition), which is often ignored by the main stream of literatures. The rest steps are apparently illustrated in the flow chart (Figure 11).
Conclusion
Hereby, we discovered novel drug candidates against leukemia, which will lead to new and better therapeutics. Leukemia is a serious cancer threatening all ages of people globally. To date, no method that can prevent leukemia has been testified by medical evidence (NCI, 2024) [42]. Thus, a good therapy to treat leukemia is crucial to save patients’ lives. As reviewed in this article, almost all current therapies have disadvantages, including negative effects. Better therapeutics with extraordinary specificity are in high demand to overcome the shortcomings of current therapies. We identified a lead compound. Through in-depth analysis, we revealed 13 novel molecules, who could be promising drug candidates for future structure-based drug design/development (SBDD), which will be performed to select the best candidate(s) with the best specificity. Furthermore, we propose a representative workflow, named Sun’s Flow Chart in Drug Discovery, to standardize drug development process. While “all roads lead to Rome”, the Sun’s Flow Chart is one of most simple, rational and straightforward approaches.
Acknowledgements
W.S. would thank the technical support and helpful discussions from Dr. Ken Sasaki, a former Chief Scientific Officer of SGS, a worldly industrial leader for 150 years since 1878.
References
- Taylor HG, Butler WM, Rhoads J, Karcher DS and Detrick-Hooks B, et al. (1982) Prolymphocytic leukemia: treatment with combination chemotherapy to include doxorubicin. Cancer 49(8): 1524-1529.
- Bern MM, Wallach SR, Arkin CF, Lokich JJ, Huberman MS, et al. (1987) Etoposide in combination with cytarabine, doxorubicin, and 6-thioguanine for treatment of acute nonlymphoblastic leukemia in a protocol adjusted for age. Cancer Treat Rep 71(2): 201-203.
- Pan XQ, Zheng X, Shi G, Wang H, Ratnam M, et al. (2002) Strategy for the treatment of acute myelogenous leukemia based on folate receptor beta-targeted liposomal doxorubicin combined with receptor induction using all-trans retinoic acid. Blood 100(2): 594-602.
- Reimer RR, Hoover R, Fraumeni JF Jr, Young RC (1977) Acute leukemia after alkylating-agent therapy of ovarian cancer. N Engl J Med 297(4): 177-181.
- Kempin S, Lee BJ, Thaler HT, Koziner B, Hecht S, et al. (1982) Combination chemotherapy of advanced chronic lymphocytic leukemia: the M-2 protocol (vincristine, BCNU, cyclophosphamide, melphalan, and prednisone). Blood 60(5): 1110-1121.
- Jordan MA, Himes RH, Wilson L (1985) Comparison of the Effects of Vinblastine, Vincristine, Vindesine, and Vinepidine on Microtubule Dynamics and Cell Proliferation in Vitro. Cancer Res 45(6): 2741-2747.
- Relling MV, Yanishevski Y, Nemec J, Evans WE, Boyett JM, et al. (1998) Etoposide and antimetabolite pharmacology in patients who develop secondary acute myeloid leukemia. Leukemia 12(3): 346-352.
- Deininger M, Buchdunger E, Druker B J (2005) The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood 105(7) 2640-2653.
- Blagosklonny MV (1999) Drug-resistance enables selective killing of resistant leukemia cells: exploiting of drug resistance instead of reversal. Leukemia 13(12): 2031-2035.
- Nucifora G, Larson R, Rowley J (1993) Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood 82(3): 712-715.
- Licht J, Chomienne C, Goy A, Chen A, Scott A, et al. (1995) Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17). Blood 85(4): 1083-1094.
- Huntly BJP, Reid AG, Bench AJ, Campbell LJ, Telford N, et al. (2001) Deletions of the derivative chromosome 9 occur at the time of the Philadelphia translocation and provide a powerful and independent prognostic indicator in chronic myeloid leukemia. 10.1182/blood V98.6.1732 Blood 98(6): 1732-1738.
- Mistry AR, Felix CA, Whitmarsh RJ, Mason A, Reiter A, et al. (2005) DNA topoisomerase II in therapy related acute promyelocytic leukemia. N Engl J Med 352(15): 1529-1538.
- Felix CA, Lange BJ, Hosler MR, Fertala J, Bjornsti MA, et al. (1995) Chromosome Band 11q23 Translocation Breakpoints Are DNA Topoisomerase II Cleavage Sites. Cancer Res 55(19): 4287-4292.
- Kanoe H, Nakayama T, Hosaka T, Murakami H, Yamamoto H, et al. (1999) Characteristics of genomic breakpoints in TLS-CHOP translocations in liposarcomas suggest the involvement of Translin and topoisomerase II in the process of translocation. Oncogene 18(3): 721-729.
- Wang JC (1996) DNA topoisomerases. Annu Rev Biochem 65(1): 635-692.
- Schneider YJ, Baurain R, Zenebergh A, Trouet A (1979) DNA-binding parameters of daunorubicin and doxorubicin in the conditions used for studying the interaction of anthracycline-DNA complexes with cells in vitro. Cancer Chemother Pharmacol 2(1):7-10.
- Larsen AK, Escargueil AE, Skladanowski (2003) A Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol Ther 99(2)167-181.
- Gormley NA, Orphanides G, Meyer A, Cullis PM, Maxwell A, et al. (1996) he interaction of coumarin antibiotics with fragments of DNA gyrase B protein. Biochemistry 35(15): 5083-5092.
- Eder JP, Teicher BA, Holden SA, Cathcart KN, Schnipper LE, et al. (1987) Novobiocin enhances alkylating agent cytotoxicity and DNA interstrand crosslinks in a murine model. J Clin Invest 79(5): 1524-1528.
- Rappa G, Lorico A, Sartorelli AC (1993) Reversal of etoposide resistance in non-P-glycoprotein expressing multidrug resistant tumor cell lines by novobiocin. Cancer Res 53(22): 5487-5493.
- Gottesfeld JM (1986) Novobiocin inhibits RNA polymerase III transcription in vitro by a mechanism distinct from DNA topoisomerase II. Nucleic Acids Res 14(5): 2075-2088.
- Drake FH, Hofmann GA, Mong SM, Bartus JO, Hertzberg RP, et al. (1989) In vitro and intracellular inhibition of topoisomerase II by the antitumor agent merbarone. Cancer Res 49(10): 2578-2583.
- Fortune JM, Osheroff N (1998) Merbarone inhibits the catalytic activity of human topoisomerase IIalpha by blocking DNA cleavage. J Biol Chem 273(28): 17643-17650.
- Wang L, Roy SK, Eastmond DA (2007) Differential cell cycle-specificity for chromosomal damage induced by merbarone and etoposide in V79 cells. Mutat Res 616(1-2) 70-82.
- Glover A, Chun HG, Kleinman LM, Cooney DA, Plowman J, et al. (1987) Merbarone: an antitumor agent entering clinical trials. Invest New Drugs 5(2): 137-143.
- Kraut EH, Bendetti J, Balcerzak SP, Doroshow JH (1992) Phase II trial of membrane in soft tissue sarcoma. A Southwest Oncology Group study. Invest New Drugs 10(4): 347-349.
- Malik UR, Dutcher JP, Caliendo G, Lasala P, Mitnick R, et al (1997) Phase II trial of membrane in patients with malignant brain tumors. Med Oncol 14(3-4): 159-162.
- Wentland MP, Lesher GY, Reuman M, Gruett MD, Singh B, et al. (1993) A. Mammalian topoisomerase II inhibitory activity of 1-cyclopropyl-6,8- difluoro-1,4-dihydro-7-(2,6-dimethyl-4-pyridinyl)-4-oxo-3-quinolinecarb oxylic acid and related de-rivatives. J Med Chem 36(19): 2801-2809.
- Permana PA, Snapka RM, Shen LL, Chu DT, Clement JJ, et al. (1994) Quinobenoxazines: a class of novel antitumor quinolones and potent mammalian DNA topoisomerase II catalytic inhibitors. Biochemistry 33(37): 11333-11339.
- Coughlin SA, Danz DW, Robinson RG, Klingbeil KM, Wentl MP, et al. (1995) Mechanism of action and antitumor activity of (S)-10-(2,6-dimethyl-4-pyridinyl)-9-fluoro-3-methyl-7-oxo-2,3-dihydro-7 H- pyri-dol[1,2,3-de]-[1,4]benzothiazine-6-carboxylic acid (WIN 58161). Biochem Pharmacol 50(1): 111-122.
- Tabarrini O, Cecchetti V, Fravolini A, Nocentini G, Barzi A, et al. (1999) Design and synthesis of modified quinolones as antitumoral acridones. J Med Chem 42(12): 2136-2144.
- Reuman M, Daum SJ, Singh B, Wentland MP, Carabateas PM, et al. (1989) Synthesis and antibacterial activity of some novel 1-substituted-7-pyridinyl-1,4-dihydro-4-oxoquinoline-3carboxylic acids. Twenty-nineth Infer science Conference on Antimicrobial Agents and Chemotherapy: Houston, TX.
- Chu DT, Hallas R, Clement JJ, Alder J, McDonald E, et al. (1992) Synthesis and antitumor activities of quinolone antineoplastic agents. Drugs Exp Clin Res 18(17): 275-282.
- Lesher GY (1989) United States Patent 4,839,355.
- Vree TB, Wijnands WJA, Guelen PJM, Baars AM, Hekster YA, et al. (1986) Pharmacokinetics: metabolism and renal excretion of quinolones in man. Pharmacy World & Science 8(1) 29-34.
- Stahle HA (2000) historical perspective: development of clonidine. Best Practice & Research in Clinical Anesthesiology 14(2) 237-246.
- Krygowski TM, Szatylowicz H, Zachara JE (2005) How H-bonding Modifies Molecular Structure and pi-Electron Delocalization in the Ring of Pyridine/Pyridinium Derivatives Involved in H-Bond Complexation. J. Org. Chem 70(22): 8859-8865.
- Silinskas KC, Okey AB (1975) Protection by 1,1,1-trichloro-2,2-bis(p-chlorophenyl) ethane (DDT) against mammary tumors and leukemia during prolonged feeding of 7,12-dimethylbenz(a)anthracene to female rats. J Natl Cancer Inst 55(3): 653-657.
- Lin JH Lu AYH (1997) Role of Pharmacokinetics and Metabolism in Drug Discovery and Development. Pharmacol Rev 49(4): 403-449.
- Sun W (2024) Designing Effective Drugs Against Covid: Overcoming the Obstacles. Drug Des Int Prop Int J 4: 477-482.
- National Cancer Institute (NCI). 2024. Leukemia—Patient Version. Retrieved from NCI, Cancer Types.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...