
Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4692
Short Communication(ISSN: 2637-4692) 
Association of Prx Gene Expression with the Gingival Phenotype. Volume 4 - Issue 4
Mary S. Kalamaki1, Aikaterini El Doufexi*2, Maria Zempila3 and Antonis Konstantinidis4
- 1Associate Professor, Department of Science and Technology, American College of Thessaloniki, Greece
- *2Associate Faculty, Department of Preventive Dentistry, Periodontology and Dental Implant Biology, Aristotle University of Thessaloniki, Greece
- 3Graduate Student, Department of Preventive Dentistry, Periodontology and Dental Implant Biology, Aristotle University of Thessaloniki, GreeceGreece
- 4Professor, Department of Preventive Dentistry, Periodontology and Dental Implant Biology, Aristotle University of Thessaloniki, GreeceGreeceGreece
Received: March 18, 2021 Published: April 5, 2021
Corresponding author: Aikaterini El Doufexi, Associate Faculty, Department of Preventive Dentistry, Periodontology and Dental Implant Biology, Aristotle University of Thessaloniki, Greece
DOI: 10.32474/MADOHC.2021.04.000195
Keywords
Gingival phenotype; Prx; Periodontal surgery; Transcription factor
Introduction
In addition to their role in development Prx genes have important roles in cell proliferation and migration [1]. Smooth muscle cells transfected with rat Prx1 showed increased proliferation [1]. Moreover, treatment of fibroblasts in vitro with morpholino oligos designed to specifically inhibit Prx1 and Prx2 protein expression resulted in downregulation of Tenascin-C expression and reduction in cell migration towards fibronectin [1]. Specifically, the healing process after surgical manipulation in the oral mucosa, is determined in a high degree by the gingival phenotype. Assuming that the gingival phenotype might be regulated by gene expression, the aim of our study was to investigate if there is an association between the presence and the levels of Prx1 and Prx2 mRNA and the phenotype of human gingivae.
Materials and Methods
Subjects included in the study: Twenty two patients in need
of crown lengthening were included in the study, ten with thick and
twelve with thin gingival phenotype. The assessment of the gingival
phenotype was done based on the clinical characteristics of the
gingiva.
Surgical Procedures: All surgeries were performed by
postgraduate students at the Department of Periodontology
and Biology of Dental Implants at the Aristotle University of
Thessaloniki. All patients were required to sign a consent form
before.
Tissue samples: Connective tissue fragments 1mm x 1mm were
collected from the buccal flap during the surgical procedure and
immediately placed in sterile tubes containing 1mL of RNA later
(Sigma). Samples were maintained in RNA later at -80°C until
further processing.
RNA isolation-Reverse Transcription: Total RNA was
isolated from tissue samples using the NucleoSpin RNA Kit
(Macherey Nagel) following the manufacturer’s protocol. The
concentration and quality of RNA in each sample was determined
using a SmartSpec Plus spectrophotometer (Biorad). 200ng of
RNA were reversed transcribed to cDNA using the PrimeScript II
1st strand cDNA synthesis kit (Takara) using oligo-dT primer. cDNA
samples were stored at -20°C until further processing.
PCR Analysis for Prx1 and Prx2 mRNA Expression: The
presence and levels of Prx1 and Prx2 mRNA expression were
examined by semiquantitative PCR analysis. Two microliters
of the cDNA were directly used as a template for the PCR in a
total reaction volume of 25μL. The reaction mixture contained
12.5μL OneTaq Hot Start 2x Master Mix with standard buffer
(New England Biolabs), 1μL of each primer (10μM), 2μL cDNA
template, and PCR grade water. For detecting the presence of
Prx1 primers Prx1F 5’-TCCCTCCTCAAATCCTAC-3’ and Prx1R
5’-ACTATATTCCTTGGCCTTC-3’ [2] were used.
For Prx2 detection primers Prx2F 5’-AGCACAGTGCCACCCTACA-3’
and Prx2R 5’-CTTGGCAGGCTCTTCCACC-3’ [2]. Expression of Gadph was used as an endogenous control. The primers for Gadph
detection were: Forward 5’-TGG TAT CGT GGA AGG ACT CATGAC-3’
and Reverse: 5′-ATG CCA GTG AGC TTC CCGTTC AG-3′ [3].
PCR conditions were as follows: initial denaturation for 5
minutes at 94°C, then 25 cycles of denaturation at 94°C for 30s,
annealing at 48°C for Prx1, 52°C for Prx2 and 48°C for Gadph for
30s, extension for 45s at 68°C and final extension at 68°C for 5 min.
PCR products were visualized in a 2% agarose gel using Midori
green Advance (Nippon Genetics) DNA/RNA stain and a FastGene
LED Transilluminator (Nippon). Semi-quantification of the PCR
products was done by using the ImageJ (https://imagej.nih.gov/)
analysis program. All experiments were repeated twice to ensure
statistical accuracy.
Statistical analysis: The differences between thin and thick
phenotypes were analyzed using t-test. Inter-group differences
were analyzed by t-test and Mann-Whitney test for independent
samples. A p- value <0.05 was considered statistically significant.
Results
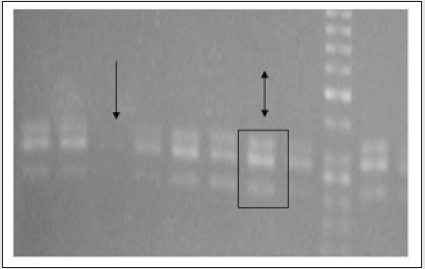
PCR Analysis for Prx1 mRNA Expression: PCR analysis showed Prx1 mRNA expression in higher levels in thick gingival phenotypes compared to thin. In addition, both Prx1a and Prx1b alternatively spliced isoforms were detected and Prx1a was more abundant in most of the samples compared to Prx1b. In thin gingival phenotypes Prx1a was either expressed in different levels or completely absent, fact that indicates that thin gingival phenotype might be regulated by other genes’ expression (Figure 1).
Figure 1: Paired related homeobox 1a and b gene isoform expression. The single arrow indicates the absence of Paired related homeobox 1 Paired related homeobox expression in a sample with thick gingival phenotype and the double arrow indicates the presence and abundance of both isoforms of Paired related homeobox 1 gene.
PCR Analysis for Prx2 mRNA Expression: PCR analysis showed that Prx2 mRNA expression was absent in thick gingival phenotypes except for one sample. On the contrary, Prx2 was highly expressed in all samples with thin gingival phenotype (Figure 2).
Figure 1: Paired related homeobox 2 gene expression. The single arrows indicate the high levels of Paired related homeobox 2 gene expression in samples with thin gingival phenotypes.

Discussion
Recent studies determine the gingival phenotype based only on clinical characteristics of the gingiva. Placing the periodontal probe in the gingival sulcus can give the appearance of transparency of the gingiva and this biotype is characterized as ‘thin’. If the gingiva is not transparent when placing the probe, the gingiva is characterized as ‘thick’ [4,5]. In this study we aimed to associate the expression of Prx genes with the gingival phenotype. Our results indicate that Prx2 is expressed in thin phenotypes, whereas Prx1 is expressed in thick phenotypes. It is well established that Prx genes play an important role in craniofacial development. Our study indicates that Prx genes are not only expressed in tissues of ectomesenchymal origin, but they might also play an important role in determining the gingival phenotype at a molecular level. It is interesting t that Prx2 was almost exclusively expressed in thin phenotypes and in the same samples Prx1 was absent, implying opposing roles of the two genes in adult gingival tissues. Overall, our study indicates a strong association of Prx1 and Prx2 gene expression with the gingival phenotype. It is possible that Prx gene expression can be used as a molecular screening test to define more accurately the gingival phenotype.
Acknowledgment
Apostolos S. Angelidis, D.V.M., M.P.V.M., Ph.D. Laboratory of Milk Hygiene and Technology Department of Food Hygiene and Technology School of Veterinary Medicine Aristotle University of Thessaloniki, Greece.
References
- Jones FS, Meech R, Edelman DB, Oakey RJ, Jones PL (2001) Prx1 Controls Vascular Smooth Muscle Cell Proliferation and Tenascin-C Expression and Is Upregulated with Prx2 in Pulmonary Vascular Disease. 89(2): 131-138.
- Norris RA, Scott KK, Moore CS, Stetten G, Brown CR, et al. (2000) Human PRRX1 and PRRX2 genes: Cloning, expression, genomic localization, and exclusion as disease genes for Nager syndrome. Mammalian Genome 11(11): 1000-1005.
- Ko SY, Lin SC, Wong YK, Liu CJ, Chang KW, et al. (2007) Increase of disintergin metalloprotease 10 (ADAM10) expression in oral squamous cell carcinoma. Cancer Letters 245(1-2): 33-43.
- Fischer KR, Künzlberger A, Donos N, Fickl S, Friedmann A (2018) Gingival biotype revisited-novel classification and assessment tool. Clinical Oral Investigations 22(1): 443-448.
- Fürhauser R, Mailath-Pokorny G, Haas R, Busenlechner D, Watzek G, et al. (2017) Immediate Restoration of Immediate Implants in the Esthetic Zone of the Maxilla Via the Copy-Abutment Technique: 5-Year Follow-Up of Pink Esthetic Scores. Clinical Implant Dentistry and Related Research 19(1): 28-37.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...




