Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2644-1217
Research ArticleOpen Access
Bioremediation of Cypermethrin by Laccase Producing Bacteria Volume 4 - Issue 2
Amrita G Kulkarni, Sumati Biradar, Shipa BN, Madhu B and AB Vedamurthy*
Department of Studies in Biotechnology and Microbiology, Karnataka University, India
Received: September 10, 2022 Published: September 21, 2022
*Corresponding author: AB Vedamurthy, Department of Studies in Biotechnology and Microbiology, Karnataka University, Dharwad, 580 003, India
DOI: 10.32474/OAJCAM.2022.04.000181
Abstract
Increased urbanization and industries has led to the production, accumulation and incorporation of contaminant into the ecosystems, distorting and impacting, plant, human and animal health. Several physical, chemical, and biological strategies have been adopted to eliminate these contaminants from soil and water bodies. Biotechnological processes involving microorganisms and enzymes have been widely used for this purpose; specifically, laccases, which is a versatile enzyme which catalyse oxidation reaction coupled to four- electron reduction of molecular oxygen to water. Laccases 1, 4-benzenediol oxygen oxidoreductase, are also called as “Blue Enzyme” as they are multinuclear copper containing enzymes. Laccase belongs to the superfamily of multicopper oxidases (MCOs)—a group of enzymes comprising many proteins with different substrate specificities and diverse biological functions. are broad spectrum biocatalysts, have been used to degrade several compounds, such as those that can be found in the effluents from industries and hospitals. Cypermethrin is an organophosphorus pesticide, belonging to a group of synthetic pyrethroid insecticides which are analogous of naturally occurring pyrethrins of botanical origin. It is widely used in agriculture, forestry, Horticulture, public health, and households for the protection of textiles and to check pest infestation. The present investigation is therefore carried out to study the potential of bacterial laccase in the biodegradation of Cypermethrin.
Keywords: Laccase; Bacteria; Bioremediation; Cypermethrin
Introduction
Laccase belongs to the enzyme family of multi- copper oxidases (MCOs), are classified as benzenediol oxygen reductases and are also known as Uracil oxidases and p-diphenol oxidases. The term ‘Laccase’ stems from its original identification in the exudates of the Japanese lacquer tree Rhusver nicifera described by Yoshida (1883). Just over a century later, it was characterized as being a metal-containing oxidase depicted by Bertrand (1985). Laccase can catalyse the oxidation of various organic and inorganic compounds including diphenols, polyphenols, diamines, substituted phenols, and aromatic amines with reduction of molecular oxygen to water. Laccase also oxidizes other substrates such as aromatic amines, syringaldazine, 2,2-azinobis(3-ethylbenzothiazoline6-sulphonate) and 1hydroxybenzotriazole, 2,2,6,6-tetrametylpiperidineoxy (TEMPO), violuric acid to form free radicals (Zille et al., 2003).
Laccases have been found in plants, insects, and bacteria, but are predominant in fungi. Laccase activity has been demonstrated in more than 60 fungal strains belonging to Basidiomycetes, Ascomycetes and Deuteromycetes, being documented in virtually every fungus examined for it. Laccase is a dimeric or tetrameric glycoprotein and usually contains four Copper (Cu) atoms distributed among the three binding sites.These enzymes contain 15-30% carbohydrate and have a molecule mass of 50-90kDa. These are copper containing 1, 4-benzenediol: Oxygen oxidoreductases. (Imran et al., 2008) Among all, most well-known lignin degradation enzymes have been classified from fungi, typically from those belonging to White rot fungi, Basidiomycetes and several bacterial species from the genus Azospirillum, Streptomyces and Bacillus. The enzymes produced by such microorganisms mainly include Lignin peroxide (LiP), Manganese peroxide (MnP) and laccases Gassara et al. Due to its low specificity for the reducing substrate and are thermostable at high chloride concentration and wide pH range attracted increasing attention in industrial and environmental fields, such as pulp delignification, dye decolorization, degradation of pesticides and insecticides, water detoxification, biopolymer modification and biotransformation, soil bioremediation, biological degradation of phenolic pollutant compounds in environment. (Ranocha et al.).
In fungi, they play an important role in pigment formation, detoxification and lignin degradation. In bacteria, laccases are having their role in melanin production, detoxification of copper, spore coat resistant and are involved in morphogenesis. The bacterial laccases are having many advantages over classical fungal laccases due to their highly efficient expression, higher thermostability. Cypermethrin belongs to a group of synthetic pyrethroid insecticides which are analogous of naturally occurring pyrethrins of botanical origin. It is widely used in agriculture, forestry, Horticulture, public health and households for the protection of textiles and to check pest infestation (Zhang et al., [1]). Cypermethrin is used against pests in cotton and vegetable crops as a replacement of organophosphorus pesticide (Lin et al., [2]). Halflife of cypermethrin in soil varies from 4 to 65 days. Cypermethrin is an environment pollutant because of its widespread use, toxicity and persistence which may lead to serious damage to non-target organisms and various ecosystems. Therefore, it is necessary to develop a rapid and efficient process to eliminate or minimize the concentration of this pesticide in the environment. Variety of physical and chemical methods are available to treat the contaminants with hazardous chemicals in the soil/ ground water but most of the methods do not actually destroy the hazardous compounds rather help them in binding to the matrix or convert them from one phase to another. Biological treatment of chemically contaminated soil is simple, eco-friendly and economic and involves the transformation of complex or simple chemical compounds into non-hazardous forms. For biodegradation, target pesticide acts as a sole source of carbon and energy for the growth of microorganisms which utilize toxic compounds by producing desired enzymes. The specificity of these enzymes involving xenobiotic compounds differs from one microorganism to another. Several microbes involved in biodegradation of β-cypermethrin have been identified in recent years, such as Serratia sp., Ochrobactrum lupini DG-S- 01 and Pseudomonas aeruginosa CH (Chen et al., 2011). Three genes, i.e., Estp, pytH, and PytZ, encoding pyrethroid degrading hydrolases from Klebsiella sp. ZD112, Sphingobium sp., and Ochrobactrumanthropi YZ-1, respectively have been identified by various authors Wu et al., [3].
Use of pesticide-degrading microbial systems for the removal of pollutants from the contaminated systems requires the understanding of ecological, physiological, and biochemical mechanisms of the degrading organisms. The present study was aimed to isolation, purification characterization of bacterial Laccase and degradation of Cypermethrin. Therefore, the current investigation focuses on Isolation and screening of laccase producing bacteria from soil. Molecular characterisation of laccase producing bacteria with the potential of degrading Cypermethrin
Materials And Methods
Sample collection
a) Soil sample was collected from the sawmill at Lakkamanahalli, Dharwad, Karnataka, India. The soil was collected from the surface to a depth using the sterile spatulas and transported to the laboratory and stored at -200C.
Isolation and screening of laccase producing bacteria from Saw dust
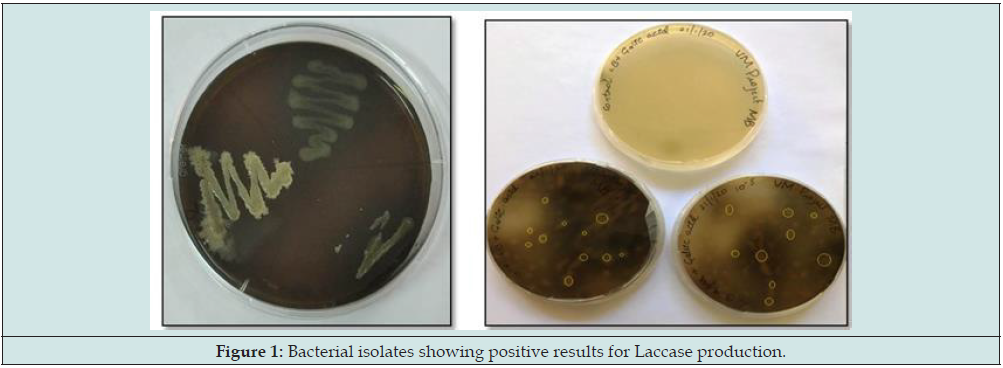
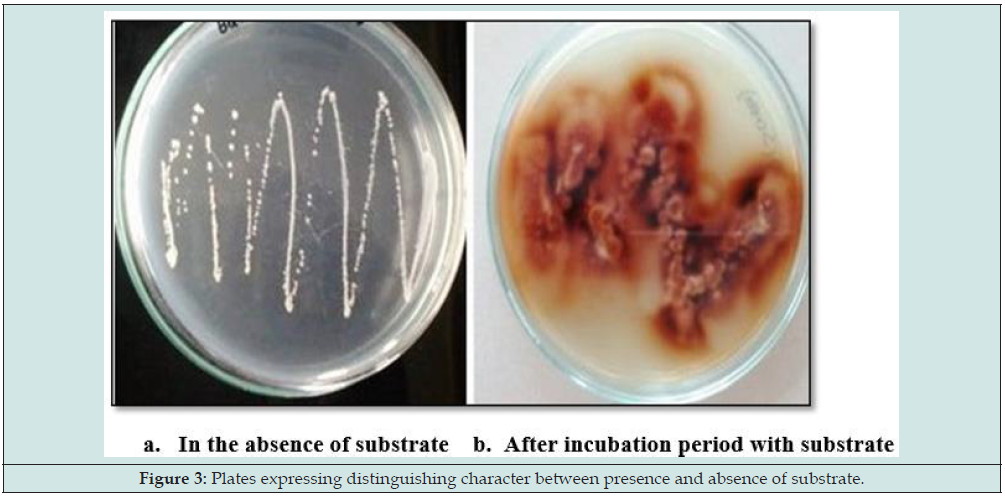
a. The isolation was based on the serial dilution technique. In this 1g of saw dust was added to a tube containing 9ml of saline/distilled water (10-1) and diluted up to 10-7. From the above dilutions, 0.1ml of suspension was spread on nutrient agar plates (NA) containing 0.2% guaiacol and 0.02% Gallic acid. The plates were incubated at 370C for 24 hrs. The plates expressing brown coloured zone were selected as positive colonies, depicts the production of laccase enzyme. The positive colonies were subcultured for further process. The laccase activity was detected in the isolates and named accordingly as Isolate1, Isolate2, Isolate3, Isolate4, and Isolate5.
Determination of Laccase Activity
I. Laccase activity was measured by monitoring the oxidation of 2mM guaiacol and gallic acid buffered with acetate buffer (pH 4.5) at 420 nm for 1minute. The reaction mixture contained 1ml of 2mM guaiacol, 1ml culture filtrate, and 3ml of acetate buffer (pH 4.5). One unit of enzyme activity was defined as the amount of enzyme that oxidized 2μmol of substrate per minute. The enzyme activity was expressed in U/ml and the highest enzyme activity was detected in the supernatant. The effect of substrate concentration on laccase production using the 5 isolates Isolate 1, Isolate 2, Isolate 3, Isolate 4, Isolate 5 showed the maximum enzyme activity of 1.073U/ml, 1.263U/ml, 1.590U/ml, 1.631U/ml, and 1.370U/ml respectively.
Partial Purification of Laccase
The supernatant obtained after cold centrifugation was concentrated by fractional precipitation up to 80% ammonium sulphate saturation. Flasks were re-suspended in 100ml of 50mM glycine NaOH buffer (pH 9.0) and centrifuged at 4,000rpm for 10minutes. After centrifugation, the sample having maximum enzyme activity was extensively dialyzed against acetate buffer. The experiments were conducted at low temperature (-40C).
Optimization of Process Parameters for laccase activity
Various process parameters that influence the enzyme production during SSF were optimized overwide range. The strategy adopted for standardization of process parameters was to evaluate the effect of an individual parameter and to incorporate it at standardized level before standardizing the next parameter.
Effect of Incubation Period on Laccase Activity
Incubation period is an important factor for the production of enzyme. 50 mL of sample was taken in 250 mL Erlenmeyer flasks. The flasks were sterilized, cooled to room temperature, and inoculated with 3.5 × 106 CFU. Incubated at different time intervals, such as, 0, 24, 48, 72, 96, 120, and 144 h. The contents of the flasks were centrifuged at 10000 rpm for 10 min at 4°C and the supernatant was used to assay the enzyme activity at 420 nm.
Effect of Temperature on Laccase Activity
Environmental temperature is a factor to which the biomass is an inescapable subject, since cell temperature must become equal to the temperature of culture medium. Temperature affects the rate of cell reaction, nature of metabolism, nutritional requirement, and the biomass concentration. 50 mL of sample was taken in 250 mL Erlenmeyer flasks. The flasks were sterilized, cooled to room temperature, and inoculated. The thermostability of partially purified laccase was analysed at different temperatures such as, 50, 60, 70, 80, 90, and 100°C. At each temperature, 40 μL of enzyme along with guaiacol and gallic acid was incubated for 10 min. The contents of the flasks were centrifuged at 10000 rpm for 10 min at 4°C and the supernatant was used to assay the enzyme activity at 420 nm.
Effect of pH on Laccase Activity
The influence of hydrogen ions on biological activities is related to their hydrogen ion concentration on enzyme activity. 50 mL of sample was taken in 250 mL Erlenmeyer flasks and pH was adjusted for each flask from 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, to 8.0, respectively by adding 1N HCl and 1 N NaOH. The flasks were sterilized, cooled to room temperature, and inoculated with 3.5 × 106 CFU. Incubated at optimized incubation period (96 h) and temperatures. Three different buffers (50 mM) were used for pH ranging from 4 to 11. Acetate buffer was used for pH 4 to 6. Phosphate buffer was used for pH 6 to 8 and glycine-NaOH buffer was used for pH 8 to 11. Forty μL of enzyme, guaiacol, gallic acid and respective buffers were incubated for 3h at optimized temperature of 70°C. The contents of the flasks were centrifuged at 10000 rpm for 10 min at 4°C and the supernatant was used to assay the enzyme activity at 420 nm.
Determination of Molecular Weight
The molecular weight of laccase was determined using Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) using 12% polyacrylamide gel and the bands were visualized with Coomassie Brilliant Blue R (Lammelliet al., 2000).
Biodegradation of Cypermethrin by Laccase Producing Bacteria
Enrichment and isolation of pesticide degrading bacterial strains was carried out in MM (Minimal Medium) by using enrichment culture technique. Bacterial colonies with different morphologies appeared on the plates were picked, purified and preserved for further use. One bacterial isolate showing maximum tolerance for cypermethrin was selected for further study. Cypermethrin-degrading bacterial Isolate 4, grown on minimal medium at 37°C for 24 h was selected and identified on the basis of morphological, biochemical, and molecular characters. Genomic DNA of the bacterial strain was extracted (Bazzicalupo et al., [4]).
To test cypermethrin biodegradation under laboratory condition bacterial isolates positive for laccase were incubated in 50 mL minimal broth containing 20 ppm cypermethrin under aseptic conditions and incubated at 37 °C. Samples were withdrawn everyday of incubation and residual pesticide was quantified by GC after extraction (Gangola et.al.,[5]). Three critical factors and their optimal ranges selected in this experiment for the analysis of cypermethrin biodegradation are, inoculum size (5, 10 and 15 mL), cypermethrin concentration (10, 20 and 30 ppm). The experiment was conducted in Minimal media for 24hours, 48hours and 72hours. Supernatant was collected by centrifuging at 24, 48 and 72 hrs of bacterial growth wherein, uninoculated medium served as control. Two microlitre of sample was centrifuged at 10,000 rpm for 10 min. Supernatant (1 ml) was transferred to Buchner funnel and mixed with sodium sulphate (1 g) and hexane (1 ml). After formation of two separate layers in separating funnel, bottom layer was discarded, and upper layer was collected in a round bottom flask. Evaporated completely in an evaporator at room temperature. To the left out of the pesticide, 2 ml hexane was added and mixed properly. After filtration, extracted solution was collected in the Eppendorf tube and analysed spectrophotometrically at 595nm.
Results
Naturally occurring microorganisms have the ability to produce various enzymes. In recent days, most of the enzymes are important for human welfare and industry. In the present study, the bacterial strains were isolated from the soil sample from mill industry because these industries are having lignin containing waste. It was degraded by native microbes that are growing over that waste. From this, it infers those microbes which are isolated from decayed wood soil might have the ability to produce laccase enzyme. For the screening process, two substrates were used, guaiacol and Gallic acid. The result is concluded to be positive based on the observation shown as in below plates. Colonies showing brown coloured zone indicates the presence of lignin degrading activity or the presence of laccase activity. From the samples, many bacterial strains were isolated and screened; it was found that some of the bacterial strains gave positive result for laccase production. The better resultants were considered for further study (Figure 1).
Morphological Characteristics of laccase producing bacteria
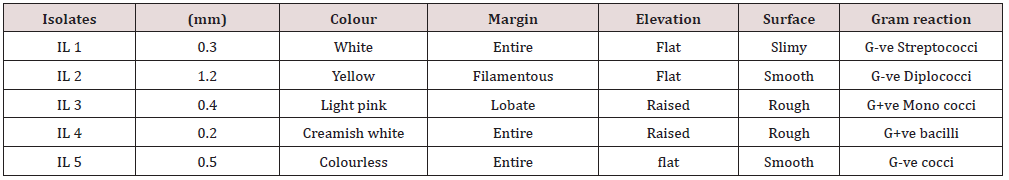
Microscopic Observation
Microscopic images of gram staining of selected strains are shown which confers the presence of gram-negative diplococci, cocci, streptococci and bacilli predominantly found in the different bacterial isolates (Figure 2).
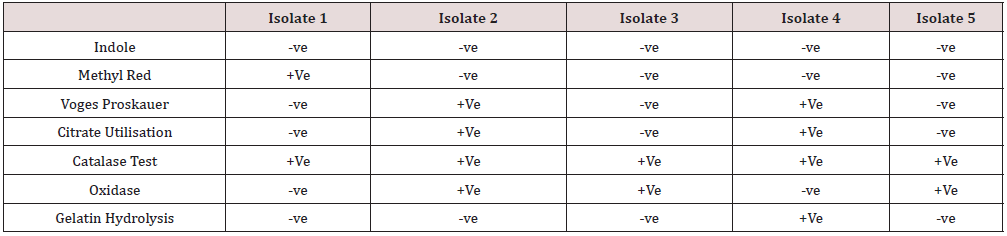
Biochemical Characteristics of Laccase Producing Bacteria
(Table 2 & Figure 3) As compared to intracellular enzymatic activity shown by these bacterial isolates, the activity is higher in the case of extracellular content by almost 80% comparatively (Graph 1).
Figure 2: Microscopic images depicting the presence of a. Gram positive streptococcib. Gram negative Diplococcic. Gram positive Coccid. Gram positive bacilli e. Gram negative cocci.
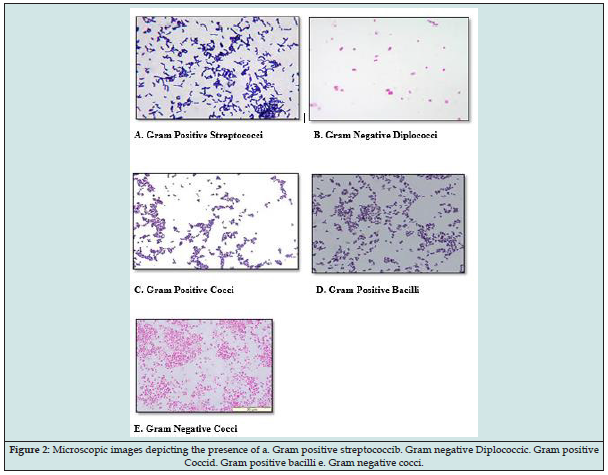
Figure 4: Comparative analysis of intracellular and extracellular production laccase enzyme by bacterial isolates.
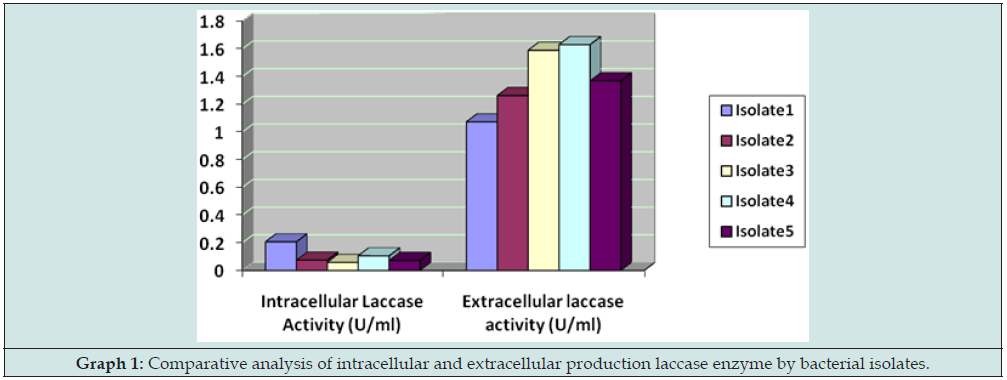
Comparative analysis of laccase activity:
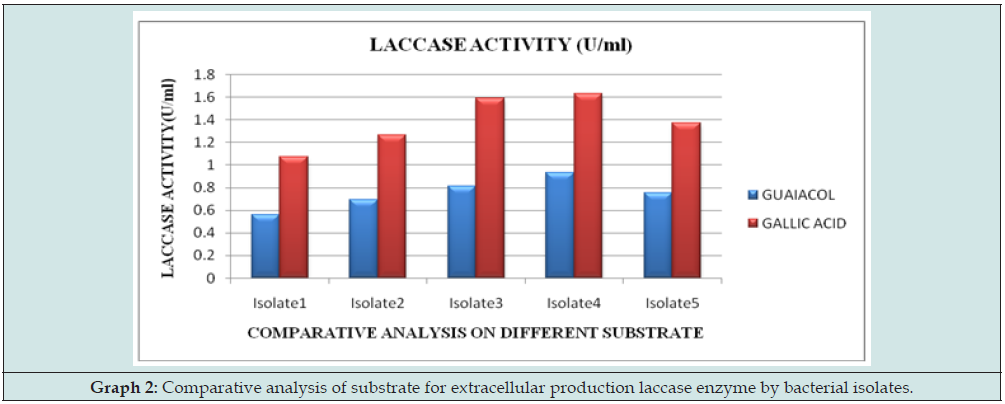
The five different bacterial isolates were subjected for mass culture and later their enzyme activity was measured as per the protocol. Optimised enzyme activity was recorded on guaiacol (0.2) than on Gallic acid comparatively. The bacterial isolates showed maximum activity extracellularly. The bacterial isolate LW01 and LB01 which has enzymes activity are being selected for further purification process. In this analysis, it was found that laccase producing bacterial isolates are more favor towards the gallic acid at 0.2% than on the guaiacol at 0.02% (Graph 2).
Figure 5: Comparative analysis of substrate for extracellular production laccase enzyme by bacterial isolates.
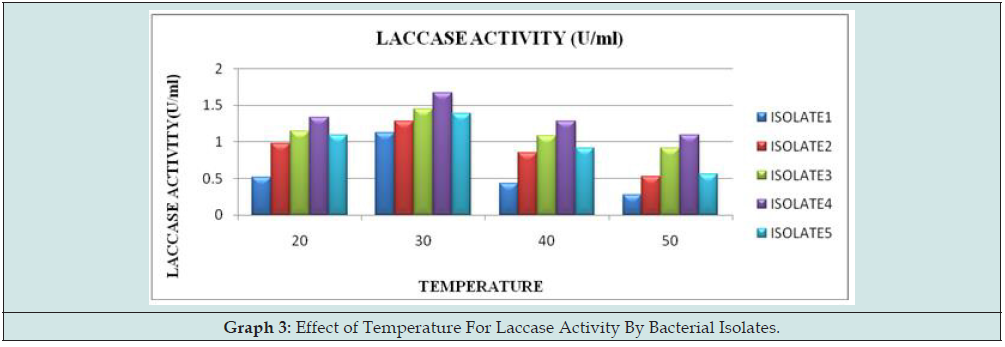
Effect of temperature on Laccase activity:
Laccase from different isolates were found to be active in the temperature range of 30 °C. Isolate 4 reported maximum activity of about 1.67U/ml at 300C (Graph 3)
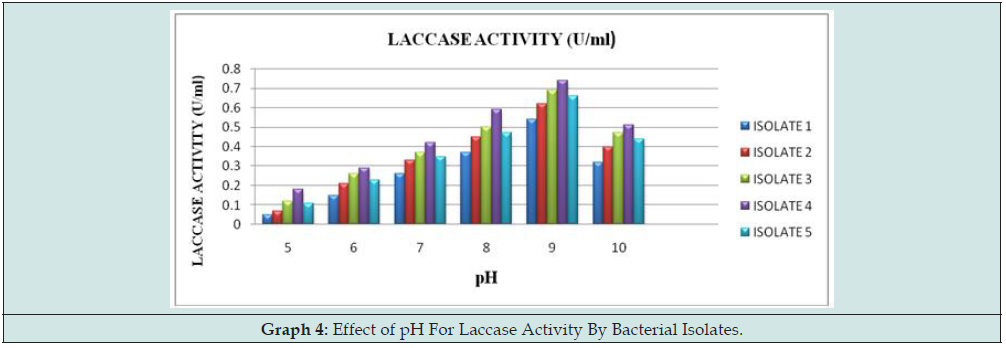
Effect of pH on enzyme activity:
Laccase from different isolates were found to be active in the pH range of 9.0. Isolate 4 reported maximum activity of about 0.74U/ ml at 9.0 (Graph 4).
Determination of molecular weight:
The molecular weights of partially purified laccase from Isolate 4 using saw dust was determined using SDS-PAGE and they were found to be approximately 52 kDa and 55 kDa for saw dust
Biodegradation of Cypermethrin by Laccase Producing Bacteria:
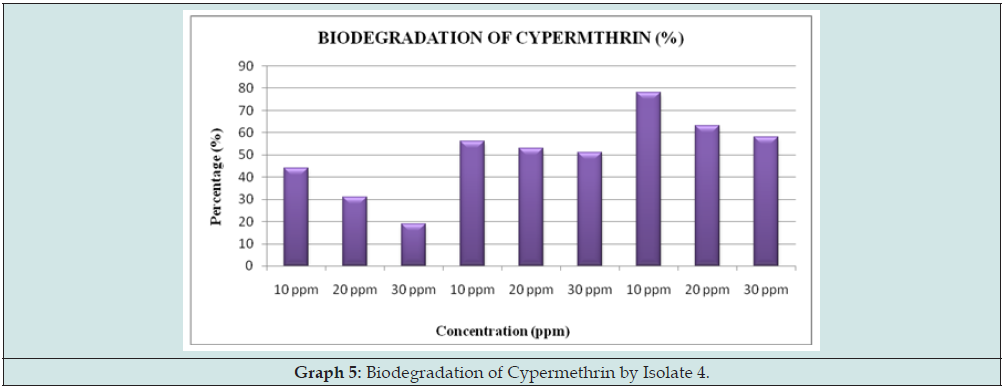
From the below observation, we can conclude that Isolate 4 is able to degrade Cypermethrin successfully at the rate of 78% at 72hours of incubation (Figure 4 & Graph 5).
From the above graph, there is successive decrease in the peak observed for 24hours, 48hours and 72hours at different concentration of 10, 20, 30 ppm. It can be inferred that Isolate 4 is successful at degrading Cypermethrin at 10ppm when compared with the other higher concentration. It also comprehends that it is actively degrading at 72hours and declines at further successive incubation period (Figure 5).
Discussion
Laccase-producing bacteria were screened from pulp and paper industry wastewater. Nutrient agar medium containing 0.5 mM of guaiacol was used. It was observed that the laccase-producing strains developed brown coloured zone from which 16 strains of Bacillus were identified. One of the isolated strains was identified as Bacillussubtilis WPI based on the results of biochemical tests and 16S rDNA sequence analysis. This strain showed laccase-like activity towards the oxidizing substrates ABTS and guaiacol. In this study guaiacol was used as the substrate of laccase activity assay. For determination of laccase activity of this isolate, guaiacol was used as a substrate of assay for the first time in this study. SDS-PAGE and Native-PAGE confirmed the presence of laccase. The laccase from B. subtilis WPI had a molecular weight of 55 kDa and was determined by SDS-PAGE (Ruijssenaars and Hartmans, [6]). A molecular weight of 56 kDa for a laccase from Bacillus halodurans and 50 kDa of from a Bacillus sp. HR03 has been determined. (Niladevi et al.,[7]).
Among the substrates, gradual increase in the enzyme activity was noted at the starting time of incubation period and the maximum enzyme activity was attained for rice bran (267 ± 2.64 U/mL) compared to wheat bran (238 ± 3.26 U/mL) at 96 h. But the production was declined at higher incubation time of 144 h. The temperature profile showed differences in the enzyme activity between rice bran and wheat bran. The maximum laccase activity was observed at 30°C (270 ± 2.78 U/mL) for rice bran when compared to wheat bran and 40°C (233 ± 4.09 U/mL) at 96 h of incubation. The enzyme activity showed a steady state of increase in their activity from pH 4.5 to 6.5, where the maximum activity was attained for rice bran (345 ± 3.14 U/mL) followed by wheat bran (265 ± 4.44 U/mL) at pH 7.0 (Chiaraluce et al., [8]). There are some reports of biodegradation of cypermethrin using microbial cultures. Degradation of cypermethrin (100 mgL−1) by Acinetobacter, Brevibacterium parabrevisup to 85% within 10 days (Akbar et al., [9]). Cypermethrin, endosulfan, imidacloprid degrading bacteria and fungi were also isolated from the rhizospheric fields (Dubey et.al., [10]) In order to obtain efficient pesticide degradative bacteria, we have screened a wide range of pesticide contaminated soil of agricultural fields, Uttarakhand, India and isolated number of bacterial and fungal isolates which can degrade cypermethrin, fipronil, imidacloprid, chlorpyrifos, carbendazim, melathion, sulfosulfuron, DDT, 2,4D and endosulfan. Few studies have suggested that Bacillus subtilis species has potential ability to degrade aromatic compounds (dye) and pesticides like cypermethrin, profenos (Arora et al. [11]).
Lignocellulosic agricultural bi-products, pearl millet (PM) and finger millet (FM) husks were used for the production of laccase using Bacillus sp. PS under solid-state fermentation (SSF).
Abiotic variables such as substrate (PM, FM) concentration (1– 5%), incubation time (24 to 96 h) and pH (5–10) were optimized using Response surface methodology (RSM) to maximize the laccase production. The predicted model showed maximum laccase activity of 402 U/mL appearing after 96 h of incubation with PM 2.0 g/L and FM 1.5 g/L at pH 7.0. Single protein band on Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) confirmed homogeneity of the laccase with a molecular weight of 50–75 kDa (Srinivasan et al., 2019), the isolate Bacillus sp. (SG2), was able to grow on MSM agar plates supplemented with 110–170 ppm cypermethrin. Degradation rate of cypermethrin increased gradually as the strain entered the logarithmic phase (4–5 days) followed by stationary phase (7–8 days). A decline in degradation rate was observed when the strain attained the death phase (12 days). More than 81 % of cypermethrin (50 ppm) was degraded by strain SG2 within 15 days in comparison to control. (Pankaj et al. [12-15]).
In this present study, isolate 4 was screened from saw dust in the Nutrient agar with 0.2 and 0.02% of Guaiacol and Gallic acid as two substrates respectively. The Isolates presented maximum laccase activity in Gallic acid. The Isolates were optimised at pH of 9.0 and temperature of 300C in the Gallic acid substrate. In this present study, Cypermethrin was degraded at 72 hours of 78%, 63% and 58% at 10, 20 and 30ppm of concentration respectively [16-19]. It was observed that the laccase-producing strains developed brown coloured zone from which 5 different organisms were isolated and identified as 16 strains of Bacillus were identified. One of the isolated strains was identified as Bacillus subtilis WPI based on the results of biochemical tests and 16S rDNA sequence analysis. This strain showed laccase-like activity towards the oxidizing substrates ABTS and guaiacol. In this study guaiacol was used as the substrate of laccase activity assay. For determination of laccase activity of this isolate, guaiacol was used as a substrate of assay for the first time in this study. SDS-PAGE and Native-PAGE confirmed the presence of laccase. The laccase from B. subtilis WPI had a molecular weight of 55 kDa and was determined by SDS-PAGL accases are the versatile enzymes which catalyse oxidation reactions coupled to fourelectron reduction of molecular oxygen to water [20-25].
In the present study, it has been proved that laccase producing bacteria are more active in the presence of Gallic acid as a substrate which shows maximum enzymatic activity which can be further studied in the applications of laccase [26-30]. It has been found through literature survey that gram negative bacteria show optimised growth in Gallic acid. Similar results have been obtained that gram-negative bacterium produce maximum laccase enzyme at 0.02 concentration of Gallic acid. From biodegradation of cypermethrin, we can conclude that isolate 4 of present study can degrade cypermethrin at 78, 63 and 58% of 10, 20 and 30ppm of concentration [31-32].
Conclusion
Laccases are produced by various sources like fungi, bacteria and insects. The use of inexpensive sources for laccase production is being explored in recent times. They have several applications because of their innate ability of oxidation of a broad range of phenolic and non-phenolic compounds. This study may also be exploited in bioremediation and cleaning practices of pesticide contaminated soil, water using the best isolates or gene/enzymes. The present work is very significant in order to degrade the xenobiotic or toxic chemicals into non-toxic end product within a short period of time through eco-friendly and economically. The present study reveals that laccase producing bacteria are more active in the presence of Gallic acid as a substrate which shows maximum enzymatic activity. Laccase enzyme has the property to act on a range of substrates and to detoxify a range of pollutants, Hence, it can be concluded that laccase producing bacteria is receiving much attention of researchers around the globe.
Acknowledgement
The authors are grateful to the Post Graduate Department of Studies in Microbiology and Biotechnology, Karnatak University Dharwad for providing the necessary facilities.
References
- Zhang C, Wang S, Yan Y (2011) Isomerization and biodegradation of beta-cypermethrin by Pseudomonas aeruginosa CH7 with biosurfactant production. Bioresource technology 102(14): 7139-7146.
- Mohammadian M, Fathi-Roudsari M, Mollania N, Badoei-Dalfard A, Khajeh K (2010) Enhanced expression of a recombinant bacterial laccase at low temperature and microaerobic conditions: Purification and biochemical characterization. J Ind MicrobiolBiotechnol 37(8): 863-869.
- Xiwen Wang, Hu Jian, Liang Yun, Zhan Huaiyu (2011) Effects of Metal Ions on Laccase Activity. Asian Journal of Chemistry 23(12): 5422-5424.
- Bazzicalupo M, Fani, R (1996) The use of RAPD for generating specific DNA probes for microorganisms. Methods Mol Biol 155-175.
- Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G (2010) Laccases: a never-ending story. Cell Mol Life Sci 67(3): 369-385.
- Shraddha, Ravi Shekher, Simran Sehgal, MohitKamthania, Ajay Kumar (2011) Laccase: Microbial Sources, Production, Purification and Potential Biotechnological Applications. Enzyme Research pp. 217861.
- Nkya TE, Akhouayri I, Kisinza W, David JP (2013) Impact of environment on mosquito response to pyrethroid insecticides: facts, evidences and prospects. Insect biochemistry and molecular biology 43(4): 407-416.
- Christopher F Thurston (1994) The structure and function of fungal laccases. Microbiology Society 140(1): 19-26.
- Akbar S, Sultan S, Kertesz M (2015) Determination of cypermethrin degradation potential of soil bacteria along with plant growth-promoting characteristics. Current microbiology 70(1): 75-84.
- Elsheikha HM, Khan (2011) NA Review of Essentials of Veterinary Parasitology. Essentials of Veterinary Parasitology Caister Academic Press, 2011:222.
- Arora PK (2012) Decolourization of 4-chloro-2-nitrophenol by a soil bacterium, Bacillus subtilis RKJ 700. PLoS One 7(12): 52012.
- Reiss R, Ihssen J, Thony-Meyer L (2011) Bacillus pumilus laccase: A heat stable enzyme with a wide substrate spectrum. BMC Bioethanol 11: 1-11.
- Alkesh I Shah, Jaydip B Jobanputra (2017) Production and Partial Purification of Laccase Produced by Bacillus species Isolated from Contaminated Soil 6(9): 17863-17869.
- Agarwal K, Chaturvedi V, Verma P (2018) Fungal laccase discovered but yet undiscovered. Bioresouces and Bioprocessing 5(1): 4.
- Alcalde M (2007) Laccases: Biological Functions, Molecular Structure and Industrial Applications. Industrial Enzymes. Springer, Dordrecht.
- Bains J, Capalash N, Sharma P (2003) Laccase from a non-melanogenic, alkalotolerantγ-proteobacterium JB isolated from industrial wastewater drained soil. Biotechnol Lett 25(14): 1155-1159.
- Chiaraluce Roberta, Muthukumarasamy Narayanan P, Jackson Beenie, Joseph Raj, Antony Sevanan, et al. (2015) Murugan, Production of Extracellular Laccase from Bacillus subtilis, MTCC 2414 Using Agroresidues as a Potential Substrate. Biochemistry Research International.
- Diana M Mate, Miguel Alcalde (2016) Laccaase: Multi-purpose biocatalyst at the forefront of biotechnology. Microb Biotechnol 10(6): 1457-1467.
- Dubey KK, Fulekar MH (2013) Rhizoremediation of pesticides: mechanism of microbial interaction in mycorrhizosphere. International Journal of Advancements in Research and Technology 2: 193-210.
- Gangola S, Sharma A, Bhatt P (2018) Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci Rep 8: 12755.
- Kaur S, Khatri M, Singh G (2017) Isolation and biochemical characterization of prokaryotic laccase. International journal of advanced technology in engineering and science 5: 637-640.
- Kumar D, Kumar A, Sondhi S, Sharma P, Gupta N (2018) An alkaline bacterial laccase for polymerization of natural precursors for hair dye synthesis. 3 Biotech 8(3): 182.
- Lin QS (2011) Biodegradation of cypermethrin by a newly isolated Actinomycetes HU-S-01 from wastewater sludge. International Journal of Environmental Science & Technology 8(1): 45-56.
- Morozova Olga, Shumakovich GP, Gorbacheva Marina, Shleev SV, Yaropolov AI, et al. (2007) Blue Laccases. Biochemistry 72(10): 1136-1150.
- Muhammad Imran, Muhammad J Asad, Saqib H Hadri, Sajid Mehmood. Department of Biochemistry, PirMehr Ali Shah Arid Agriculture University, Rawalpindi, Pakistan 2 Department of Biochemistry and Biotechnology, University of Gujrat, Pakistan.
- Negi G, Pankaj Srivastava A, Sharma A (2014) In situ biodegradation of endosulfan, imidacloprid, and carbendazim using indigenous bacterial cultures of agriculture fields of Uttarakhand, India. International Journal of Agricultural and Biological Engineering 8(9): 926-934.
- Niladevi KN, Jacob N, Prema P (2008) Evidence for a halotolerant-alkaline laccase in Streptomyces psammoticus Purification and characterization. Process Biochem 43: 654-660.
- Pankaj Sharma A, Gangola S, Khati P, Kumar G, Srivastava (2016) A Novel pathway of cypermethrin biodegradation in a Bacillus sp. strain SG2 isolated from cypermethrin-contaminated agriculture field. 3 Biotech 6(1): 45.
- Ruijssenaars HJ, HartmansS(2004) A cloned Bacillus halodurans multicopper oxidase exhibiting alkaline laccase activity. Appl Microbiol Biotechnol 65(2): 177-182.
- Srinivasan P, Selvankumar T, Kamala-Kannan S, et al. (2019) Production and purification of laccase by Bacillus sp. using millet husks and its pesticide degradation application. 3 Biotech 9(11): 396.
- Wu PC, Yu H Liu, Zhuo Y Wang, Xiao Y Zhang, He Li, et al. (2006) Molecular cloning, purification, and biochemical characterization of a novel pyrethroid-hydrolyzing esterase from Klebsiella sp. strain ZD112. Journal of agricultural and food chemistry 54(3): 836-842.
- Zhang C (2010) Biodegradation of beta-cypermethrin by two Serratia spp. with different cell surface hydrophobicity. Bioresource technology 101(10): 3423-3429.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...