Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2690-5760
Research Article(ISSN: 2690-5760) 
Isolation, Identification and Antibacterial Susceptibility of Microorganisms Collected from Live Birds Market in Sokoto Volume 4 - Issue 1
A A Ebbo* and Lawal Junaidu
- Department of Veterinary Pharmacology and Toxicology, Usmanu Danfodiyo University, Nigeria
Received:September 24, 2021; Published: October 25, 2021
Corresponding author: A A Ebbo, Department of Veterinary Pharmacology and Toxicology, Usmanu Danfodiyo University, Sokoto, Nigeria
DOI: 10.32474/JCCM.2021.04.000176
Abstract
Thickly Microbial resistance to most of the currently used antimicrobial agents is on the increase. This was studied in Sokoto by culture, isolation, identification and susceptibility testing of microorganisms from cloacal swabs in live birds. This was carried out in Veterinary Microbiology and Veterinary Pharmacology laboratories, Usmanu Danfodiyo University, Sokoto. The study was performed on 100 cloacal swab samples. The samples were collected from live birds and transferred aseptically to the laboratory. All the experimental work was done under sterile conditions. From the 100 cloacal swab samples, 97 isolates were gram negative and 3 were gram positive bacteria. Among all the isolates, 23 (23%) E. coli, 20 (20%) Salmonella spp., 22 (22%) Enterobacter spp., 16 (16%) Proteus spp., 4 (4%) Pseudomonas spp., 3 (3%) Shigella spp., Staphylococci spp., 3 (3%). In gram staining, both gram positive and grams negative bacteria were found. Isolation and identification were by observing colony morphology, grams staining and standard cultural and biochemical test. Diversified bacterial species were prevalent in cloacal swabs of broiler. However, E. coli and Salmonella spp., are of public health importance (zoonosis), so birds harboring these organisms become threat to human. It could be concluded that birds from live birds’ market in Sokoto harbor multi resistant organisms that may pass through the feces to the environment and cause potential human health hazards and can cause illness.
Keywords:Antimicrobial; Microbiology; Culture; Isolation; Identification; Susceptibility Testing
Introduction
Zoonotic pathogens pose a significant public health threat, particularly with regard to bacterial diseases [1]. Birds have long played an important role in human disease, specifically in spreading microbial pathogens [2-4]. This is likely due to several key avian traits. First, like humans, birds are found worldwide. Their ability to migrate long distances, colonize new areas, and withstand a range of environments allows for a global distribution [5-10]. Second, birds are prominent species in human-dominated habitat types. The close association of birds and humans in urban and agricultural settings facilitates zoonotic disease transfer [11- 14]. Third, birds and humans are host to some of the same bacteria species, many of which are pathogenic. While evidence for direct bacterial transmission from birds to humans is limited, several bird species have indirectly transmitted at least 12 genera of pathogenic bacteria through polluted water, ticks, and feces that lead to diarrhea, Salmonellosis, Lyme disease, and other illnesses in humans [15]. Finally, both domestic agricultural and wild birds can contaminate shared spaces and cause human infections [16]. Birdcarried diseases are, therefore, of interest because of the threat not only toward birds, but also human health [17]. Antibiotics are medicines used to prevent and treat bacterial infections, Antibiotic resistance occurs when bacteria change in response to the use of these medicines. Bacteria, not humans or animals, become antibiotic-resistant. These bacteria may infect humans and animals, and the infections they cause are harder to treat than those caused by non-resistant bacteria. Antibiotic resistance leads to higher medical costs, prolonged hospital stays, and increased mortality. The world urgently needs to change the way it prescribes and uses antibiotics. Even if new medicines are developed, without behavior change, antibiotic resistance will remain a major threat. Behavior changes must also include actions to reduce the spread of infections through vaccination, hand washing, and good food hygiene, [18]. Antibiotics have been used in animal feed for about 50 years ever since the discovery not only as an anti-microbial agent, but also as a growth-promoting agent and improvement in performance.
Tetracycline, penicillin, streptomycin and Bactrian soon began to be common additives in feed for livestock and poultry. Currently, the following antibiotics are used in livestock and poultry feed: Chlortetracycline, Procaine Penicillin, Oxytetracycline, Tylosin, Bacitracin, Neomycin Sulfate, streptomycin, Erythromycin, Linomycin, Oleandomycin, Virginamycin, and bambermycins (FDA, 1995). In addition to these antibiotics, which are of microbial origin, there are other chemically synthesized antimicrobial agents that are also sometimes used in animal feeds. These include three major classes of compounds: arsenical, nito-furan, and sulfa compounds. Arsenical compounds include arsanilic acid, 3-nitro-4-hydroxy phenylarsonic acid, and sodium arsanilate; nitro-furan compounds include furazolidone and nitro-furazone; sulfamethazine, sulfathiazole, and sulfaquinoxaline (FDA, 1995). Other chemicals are also used as antiprotozoal agents to prevent coccidiosis and histomaniasis in chickens and turkeys, (The Rise of Antibiotic- Resistant Infections” (1995). FDA Consumer, 29.) Antibiotics are used regularly in animal feed at a rate of 2 to 50 grams per ton for improved performance in the animals. The reasons include a more efficient conversion of feed to animal products, an increased growth rate and a lower morbidity/mortality rate in general. The levels of antibiotics are often increased to 50-200 grams/ton or more when specific diseases are being targeted as when the spread of a particular disease is rampant. The levels are also increased in times of stress. This increased amount is often decreased when the threat of the disease is gone, [19]. Keeping backyard poultry (chicks, chickens, ducks, ducklings, geese, and turkeys) is becoming more and more popular. People enjoy raising birds as pet, commercial poultry and having fresh eggs from their established flocks. Though keeping chickens can be fun and educational, poultry owners should be aware that chickens and other poultry used for meat and eggs can carry germs that make people sick. Some common bacterial disease share between human and birds are Campylobacteriosis (Campylobacter spp.), E. coli (Escherichia coli), and Salmonellosis (Salmonella spp.). Antibiotic resistance is rising to dangerously high levels in all parts of the world. New resistance mechanisms are emerging and spreading globally, threatening our ability to treat common infectious diseases. A growing list of infections such as pneumonia, avian tuberculosis, fowl cholera, infectious bursal disease, and foodborne diseases are becoming harder, and sometimes impossible, to treat as antibiotics become less effective. Where antibiotics can be bought for animal use without a prescription and where antibiotic are administered without confirmatory diagnosis, the emergence and spread of resistance is made worse. Similarly, in countries without standard treatment guidelines, antibiotics are often over-prescribed by health workers and veterinarians and over-used by the public. Without urgent action, we are heading for a post-antibiotic era, in which common infections and minor injuries can once again kill [18].
Infections with bacterial disease in birds are in the high increase, hence the needs for culture, isolation and identification, and also susceptibility test for proper diagnosis and treatment of this microorganism. Resistant bacteria are increasingly more prevalent, more virulent, and more diverse [19]. Their rise is a direct result of antibiotic use, regardless of their form or necessity. These antibiotic-resistant bacteria can infect both humans and animals, sometimes traveling from one to the other, both within and across national borders. The chances of antibiotic-resistant bacteria prevailing in the race for survival are in direct proportion to the volume of antibiotics used, a principle which makes it all the more critical to examine current habits and encourage rational and conservative use. The aim of the studies includes isolate and identify bacteria from sample collected at live bird’s market in Sokoto and to evaluate antibacterial susceptibility microorganism for sample collected from live bird market from Sokoto. The specific objective of the study is to isolate and identify bacteria from live birds, to use macconkey agar for selective isolation and identification of Enterobacteriaceae from cloacal swab, to inoculate on blood agar and observe for haemolysis and to classify the bacteria in gram positive and gram negative. Other objectives are to utilize specific monitoring techniques to evaluate the susceptibility of a microbe to different antibiotics, to distinguish the range of activity of an antibiotic, to recognize and define advantages and limitations of different antibiotics, to understand how to reduce inappropriate use of antibiotic and to investigate alternatives to antibiotic.
Materials And Methods
Descriptions of the Study Area
The research was carried out in the veterinary Microbiology and Pharmacology Laboratory of Faculty of Veterinary Medicine, Usmanu Danfodiyo University, Sokoto. Sokoto State is geographically located in the Northwestern part of Nigeria between longitude 11º 30º to 13º 50º East and latitude 4º to 6º 40º North. The state shares common borders with Niger Republic to the North, Kebbi State to the South, and Zamfara State to the East. The state falls in the dry Sahel surrounded by sandy Sudan type Savannah. The state, a major livestock producer lying in the arid region of the country, covers a total land area of about 32,000 square km with an estimated human population of 3,696,999 million. The state ranks second in the nation livestock population, which was estimated as 3 million cattle, 3 million sheep, 5million goats. 4600 camels and variable species of poultry including chickens, guinea fowls, ducks and turkeys (Rim, 1992). The livestock are a major source of milk, meat, and egg, and hide and skin to the inhabitants of the state, and over 75% of them are raised in the traditional free-range system living in close contact in association with human settlement.
Collection of samples
The research was carried out on different commercial live birds’ market in Sokoto. The cloacal swab samples were collected carefully from the birds. The collected samples were brought to the Microbiology laboratory of the department of microbiology under the faculty of Veterinary medicine, Usmanu Danfodiyo University Sokoto. Sterile cotton swab sticks were inserted to collect the cloacal contents of chicken for isolation and identification of organism.
Materials use in the experiment
Mueller-Hinton agar, Nutrient agar, Sheep blood, Petridish, Swab stick, Inoculating loop, Cloacal swab sample, Distilled water, Glass slide, Gram staining reagent (crystal violet, lugol iodine, 95% ethanol, safranine), Conical flask, Measuring cylinder, Microscope, Inoculation loop, Bunsen burner, Saline solution, McFarland solution, MHA plate (Mueller-Hinton Agar plates), Cotton swab, Antibiotic disks (Antibiotics of choice) ,Tooth pick, Incubator Ruler, Ethanol, Forceps, Guideline chart for interpretation of antibiotic, susceptibility, stock sample (bacteria isolate).
Isolation, Identification and Antibacterial Susceptibility
Procedure of isolation
Pure cultures of bacteria obtained by aseptically streaking representative colonies of different morphological types, which appear on the culture plates on to freshly prepared macconkey agar plates from the incubator. Discrete bacteria colonies that developed (grown) were subculture on nutrient agar slopes and incubated at 37°C for 24 hours. Pure cultures were achieved as per procedures described by OIE [20].
Procedure for identifications
Cultural, morphological and biochemical characteristics were studied in order to identify the bacteria. The cultural characteristics or colonial morphology of the bacteria grown on the macconkey and blood agar media were recorded. Gram staining was performed to study the morphology and staining characteristics of bacteria according to the technique described by Merchant and Packer (1967). Biochemical tests, such as sugar fermentation, coagulase, catalase, MR (methyl red), VP (Voges-Proskauer), Triple sugar ion test, were performed per standard methods [21].
Procedure for susceptibility testing
The susceptibility of all identified isolates to antimicrobial agents was determined by the standard Kirby-Baeur disk diffusion method. Two colonies of the isolates under test were inoculated into Mueller-Hinton broth and incubated at 37°C for 24 hours. After incubation, the turbidity of the Mueller-Hinton broth culture was adjusted to 0.5 McFarland standards using a ultra-violet spectrophotometer at 630nm wavelength. A sterile swab stick was dipped into adjusted Mueller-Hinton broth culture and inoculated onto Mueller-Hinton agar plate using spread method. The antibiotic discs were carefully placed at equal distance apart using a sterile forceps. Concentration of antibiotics disc used were: Cloxacillin 5 μg; Oxytetracycline 30μg; penicillin G 10μg; streptomycin 10μg; amoxicillin/clavulanic acid 30μg; Ciprofloxacin 5μg and gentamicin 30μg. The plates were incubated at 37°C for 24 hours. Zones of inhibition around each antibiotic disc were observed and measured in millimeter using a ruler and interpreted as susceptible, intermediate and resistance in accordance with the recommendation of Clinical and Laboratory Standards Institute [22] guidelines.
Results
Percentage Occurrence of Isolates
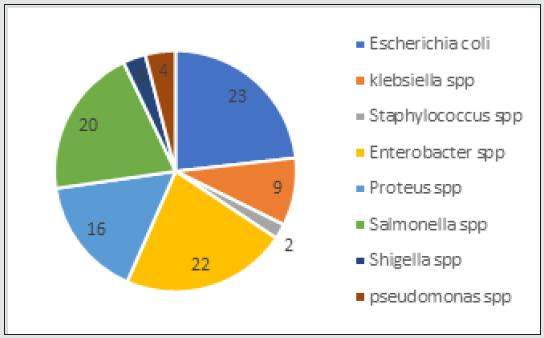
A total of one hundred (100) bacterial isolates from cloacal swab of birds in the study area were processed. Escherichia coli was the most common isolate 23(23%) and Enterobacter spp 22(22%). This was closely followed by Salmonella spp 20(20%), Proteous spp 16(16%), Klebsella spp 10(10%), Pseudomonas spp 4(4%), Shigella spp 3(3%). The least isolate was Staphylococcus spp 3(3%). This is as shown in Figure 1.
Zone of inhibition for the tested antibiotics
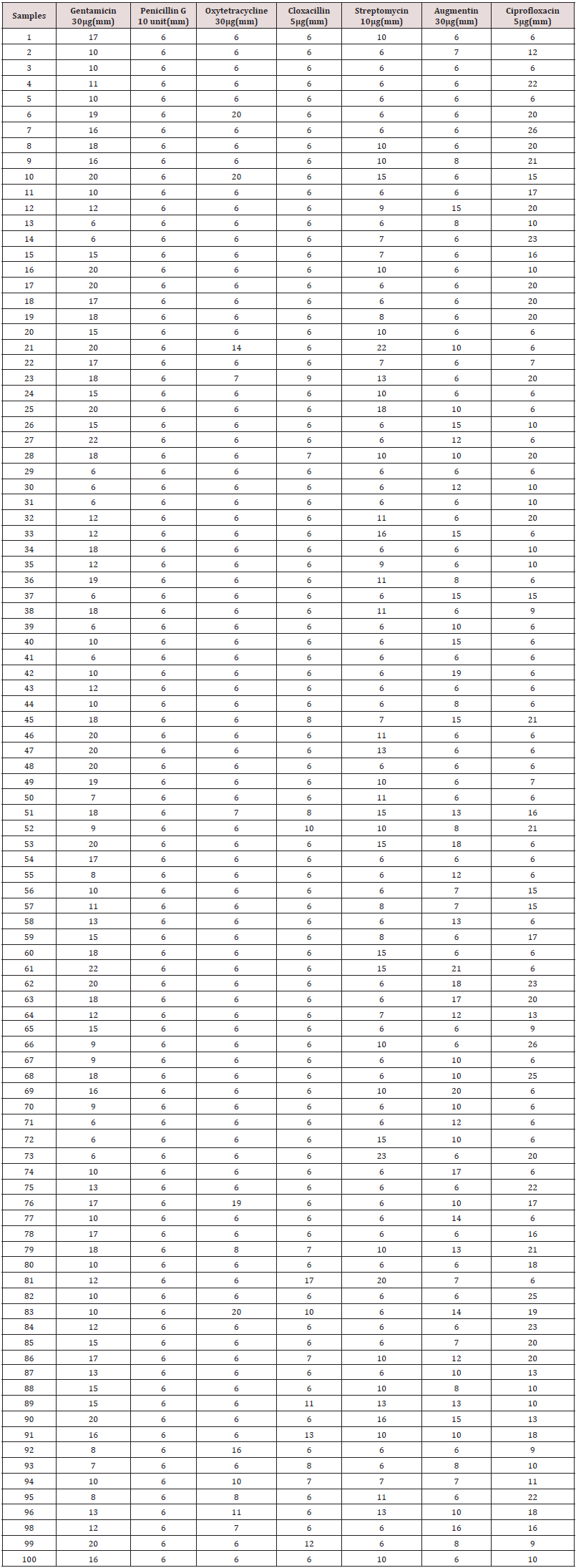
Antibiotics used for susceptibility testing were gentamicin (30 μg) had 22 mm as the highest zone of inhibition, penicillin G (10 unit) had 6 mm as the highest, Oxytetracycline (30 μg) had 20 mm as the highest zone of inhibition, cloxacillin (5 μg) had 17mm as the highest zone of inhibition, streptomycin (10 μg) had 23 mm as the highest zone of inhibition, augmentin (30 μg) had 20 mm as the highest zone of inhibition, and Ciprofloxacin (5 μg) had 26 mm as the highest zone of inhibition. The zones of inhibition were measured in millimeter (mm) as shown in Table 1.
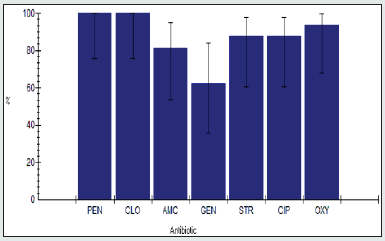
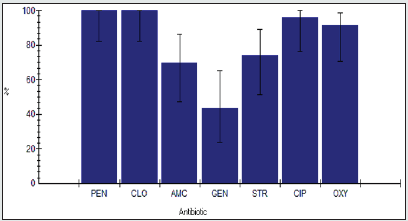
Antimicrobial Resistance among Enterobacter spp.
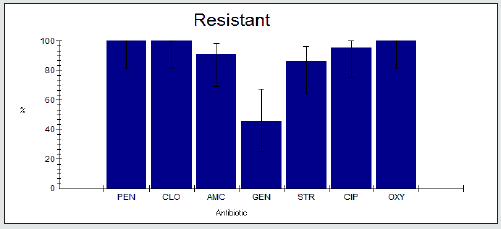
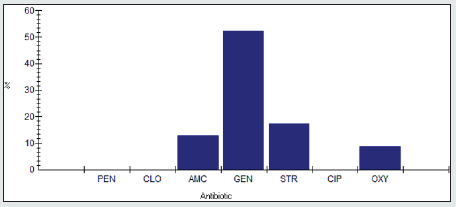
The antimicrobial resistance percentage among Enterobacter spp. to tested antibiotics are penicillin G (100%), cloxacillin (100%), amoxicillin/clavulanic acid (90.9%), gentamicin (45.5%), streptomycin (86.4%), Ciprofloxacin (95.5%) and Oxytetracycline (100%), as shown in Figure 2.
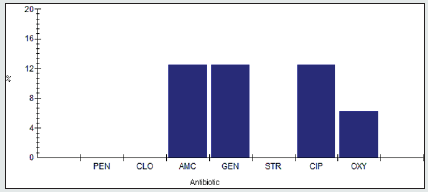
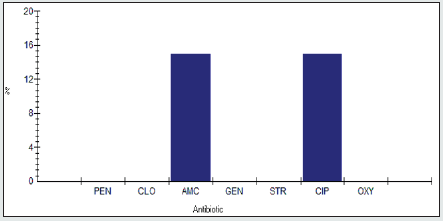
Antimicrobial Intermediate Reristance among Enterobacter spp.
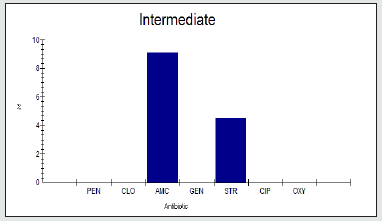
Antimicrobial intermidiate resistance percentage among Enterobacter spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (9.1%), gentamicin (0%), streptomycin (4.5%), Ciprofloxacin (0%) and Oxytetracycline (0%), as shown in Figure 3.
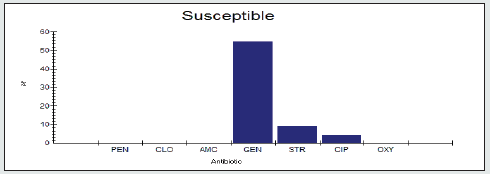
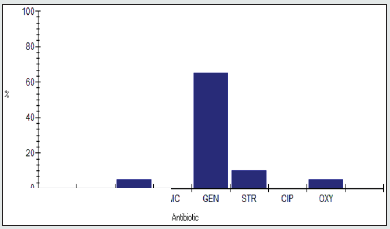
Antimicrobial Susceptibility Percentage among Enterobacter spp.
Antimicrobial susceptability percentage among Enterobacter spp to tested antibiotics are are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (0%), gentamicin (54.5%), streptomycin (9.1%), Ciprofloxacin (4.5%) and Oxytetracycline (0%), as shown in Figure 4.
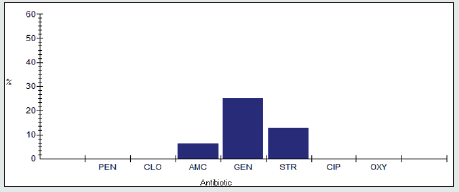
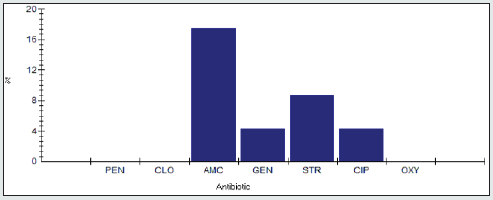
Antimicrobial Resistance Percentage among Proteus spp.
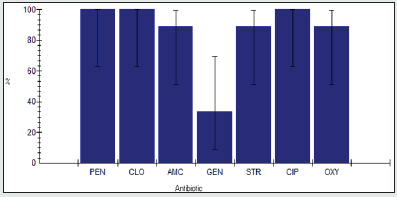
The antimicrobial resistance percentage among Enterobacter spp. to tested antibiotics are penicillin G (100%), cloxacillin (100%), amoxicillin/clavulanic acid (81.2%), gentamicin (62.5%), streptomycin (87.5%), Ciprofloxacin (87.5%) and Oxytetracycline (93.8%), as shown in Figure 5.
Antimicrobial Intermediate Percentage among Proteus spp.
Antimicrobial intermediate percentage among Proteus spp to tested antibiotics are are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (12.5%), gentamicin (12.5%), streptomycin (0%), Ciprofloxacin (12.5%) and Oxytetracycline (6.2%) as shown in Figure 6.
Antimicrobial susceptibility percentage among Proteus spp.
Antimicrobial susceptibility percentage among Proteus spp to tested antibiotics are are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (6.2%), gentamicin (25%), streptomycin (12.5%), Ciprofloxacin (0%) and Oxytetracycline (0%) as shown in Figure 7.
Antimicrobial resistance percentage among Pseudomonas spp.
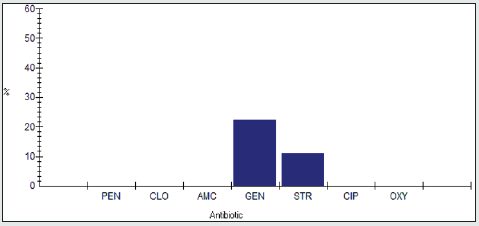
Antimicrobial resistance percentage among Pseudomonas spp to tested antibiotics are are penicillin G (100), cloxacillin (100%), amoxicillin/clavulanic acid (50%), gentamicin (0%), streptomycin (75%), Ciprofloxacin (75%) and Oxytetracycline (100%), as shown in Figure 8.
Antimicrobial intermediate percentage among Pseudomonas spp.
Antimicrobial intermediate percentage among Pseudomonas spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (0%), gentamicin (0%), streptomycin (0%), Ciprofloxacin (25%) and Oxytetracycline (0%), as shown in Figure 9.
Antimicrobial susceptibility percentage among Pseudomonas spp.
Antimicrobial susceptibility percentage among Pseudomonas spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (50%), gentamicin (100%), streptomycin (25%), Ciprofloxacin (0%) and Oxytetracycline (0%), as shown in Figure 10.
Antimicrobial resistance percentage among Salmonella spp.
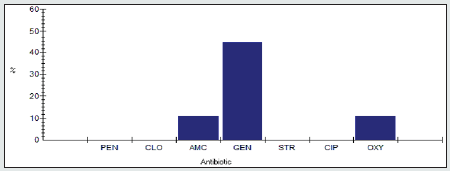
Antimicrobial resistance percentage among Salmonella spp to tested antibiotics are penicillin G (100%), cloxacillin (95%), amoxicillin/clavulanic acid (85%), gentamicin (35%), streptomycin (90%), Ciprofloxacin (85%) and Oxytetracycline (95%), as shown in Figure 11.
Antimicrobial intermediate percentage among Salmonella spp
Antimicrobial intermediate percentage among Salmonella spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (15%), gentamicin (0%), streptomycin (0%), Ciprofloxacin (15%) and Oxytetracycline (0%), as shown in Figure 12.
Antimicrobial susceptibility percentage among Salmonella spp.
Antimicrobial susceptibility percentage among Salmonella spp to tested antibiotics are penicillin G (0%), cloxacillin (5%), amoxicillin/clavulanic acid (0%), gentamicin (65%), streptomycin (10%), Ciprofloxacin (0%) and Oxytetracycline (5%), as shown in Figure 13.
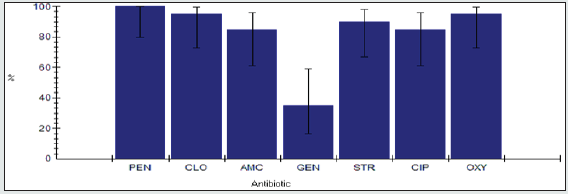
Antimicrobial resistance percentage among Escherichia coli spp.
Antimicrobial resistance percentage among Escherichia coli to tested antibiotics are penicillin G (100%), cloxacillin (100%), amoxicillin/clavulanic acid (69.6%), gentamicin (43.5%), streptomycin (73.9%), Ciprofloxacin (95.7%) and Oxytetracycline (93.1%), as shown in Figure 14.
Antimicrobial intermediate percentage among Escherichia coli spp.
Antimicrobial intermediate percentage among Escherichia coli to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/ clavulanic acid (17.4%), gentamicin (4.3%), streptomycin (8.7%), Ciprofloxacin (4.3%) and Oxytetracycline (0%), as shown in Figure 15.
<Antimicrobial susceptibility percentage among Escherichia coli spp.
Antimicrobial susceptibility percentage among Escherichia coli to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (13%), gentamicin (52.2%), streptomycin (17.4%), Ciprofloxacin (0%) and Oxytetracycline (8.7%), as shown in Figure 16.
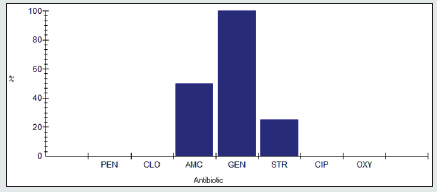
Antimicrobial resistance percentage among Klebsiella spp
Antimicrobial resistance percentage among Klebsiella spp to tested antibiotics are penicillin G (100%), cloxacillin (100%), amoxicillin/clavulanic acid (88.9%), gentamicin (33.3%), streptomycin (88.9%), Ciprofloxacin (100%) and Oxytetracycline (88.9%), as shown in Figure 17.
Antimicrobial intermediate percentage among Klebsiella spp.
Antimicrobial intermediate percentage among Klebsiella spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/ clavulanic acid (0%), gentamicin (22.2%), streptomycin (11.1%), Ciprofloxacin (0%) and Oxytetracycline (0%), as shown in Figure 18.
Antimicrobial susceptibility percentage among Klebsiella spp
Antimicrobial susceptibility percentage among Klebsiella spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/ clavulanic acid (11.1%), gentamicin (44.4%), streptomycin (0%), Ciprofloxacin (0%) and Oxytetracycline (11.1%), as shown in Figure 19.
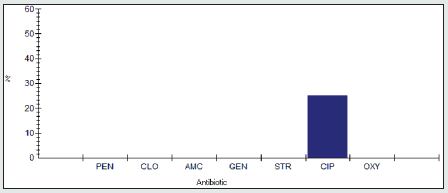
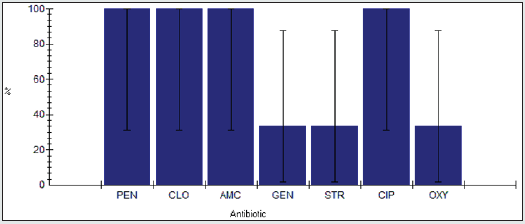
Antimicrobial resistance percentage among Shigella spp.
Antimicrobial resistance percentage among Shigella spp to tested antibiotics are penicillin G (100%), cloxacillin (100%), amoxicillin/clavulanic acid (100%), gentamicin (66.7%), streptomycin (100%), Ciprofloxacin (66.7%) and Oxytetracycline (100%), as shown in Figure 20.
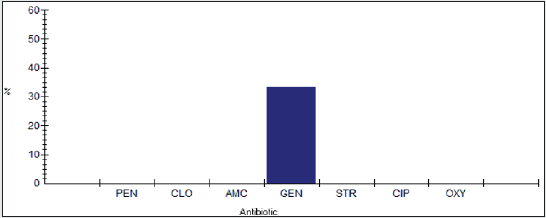
Antimicrobial intermediate percentage among Shigella spp.
Antimicrobial intermediate percentage among Shigella spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/ clavulanic acid (0%), gentamicin (0%), streptomycin (0%), Ciprofloxacin (33.3%) and Oxytetracycline (0%), as shown in Figure 21.
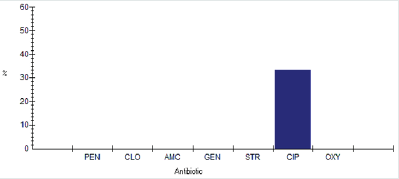
Antimicrobial susceptibility percentage among Shigella spp.
Antimicrobial susceptibility percentage among Shigella spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/ clavulanic acid (0%), gentamicin (33.3%), streptomycin (0%), Ciprofloxacin (0%) and Oxytetracycline (0%), as shown in Figure 22.
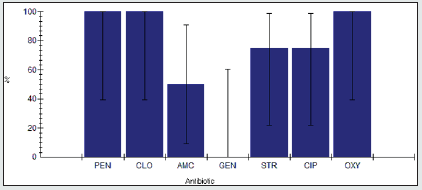
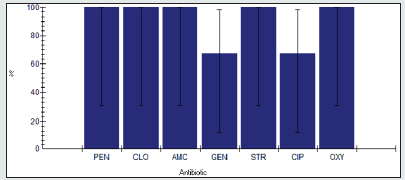
Antimicrobial resistance percentage among Staphylococcus spp.
Antimicrobial resistance percentage among Staphylococcus spp to tested antibiotics are penicillin G (100%), cloxacillin (100%), amoxicillin/clavulanic acid (100%), gentamicin (33.3%), streptomycin (33.3%), Ciprofloxacin (100%) and Oxytetracycline (33.3%), as shown in Figure 23.
Antimicrobial intermediate percentage among Staphylococcus spp.
Antimicrobial intermediate percentage among Staphylococcus spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (0%), gentamicin (0%), streptomycin (0%), Ciprofloxacin (0%) and Oxytetracycline (0%), as shown in Figure 24.
Antimicrobial susceptibility percentage among Staphylococcus spp.
Antimicrobial susceptibility percentage among Staphylococcus spp to tested antibiotics are penicillin G (0%), cloxacillin (0%), amoxicillin/clavulanic acid (0%), gentamicin (66.7%), streptomycin (66.7%), Ciprofloxacin (0%) and Oxytetracycline (66.7%), as shown in Figure 25.
Discussion, Conclusion, And Recommendations
Discussion
Of particular concern is that many of the bacteria isolated are associated with important human diseases (E coli, Salmonella, and Shigella species). The identification of the isolates obtained were generally consistent with a previous report from Ile-Ife, southwestern Nigeria [23], though a few differences were noted. In particular, Corynebacterium, Acinetobacter, and Aeromonas were reported in the Ile-Ife study and not found in the present study, while Klebsiella, and Enterobacter were encountered in this study, but not found in Ile-Ife study. These discrepancies could be due to geographical differences (Ile-Ife is in southwestern Nigeria), and it is in agreement with Ojo et al. (2012). Nonetheless, most of these organisms isolated from cloacal swab samples were E coli., Salmonella, Shigella, Enterobacter, Kebsiella, Pseudomonas Proteus, and staphylococcus species which are all gram-negative bacteria except Staphylococcus which is gram positive. This indicates higher number of gram negative than gram positive bacteria, as may be because these bacteria are normal flora of the intestinal tract of birds (Hemen et al. 2012). Interestingly, some of the bacteria encountered, including Escherichia, Staphylococcus, Pseudomonas, and Salmonella species have been isolated liberally from commercial chicken feeds in Calabar, the southernmost geopolitical zone of Nigeria (Okonko et al., 2010), suggesting that potentially, pathogenic antibiotic resistant bacteria could easily be circulated via feeding operations. This study also provides an insight into the status of antimicrobial resistance in birds in Sokoto. This part of Nigeria is of particular interest given that antimicrobials are widely used in livestock and poorly controlled and that little data is available on the effects of varying rearing practices. Furthermore, human populations in Africa are susceptible to relatively high rates of antibiotic resistant bacterial infections (Pablos-Mendez et al., 1998). Thus, a larger concern is not just the presence of potentially pathogenic bacteria resistant to antibiotics in birds, but their potential to spread through the environment and to humans, given the local farming practices. Others have observed, in both developing and industrialized nations, that a wide range of antibiotic resistant genes from agricultural operations can disseminate and persist in affected environments [24]. However, the resistance rates among isolates in this study appear to be generally (80 to 100%) higher than some of those previously reported cases in Nigeria, which ranged from 40 to 60% resistances in most instances (Ojo et al., 2012). The higher percentage resistance observed in this study may be due to the fact that it is not only laying hens that were studied but also poor regulation of antibiotic usage. [25] reported that bacterial infections occur less frequently in layers than in young broilers and consequently less antibiotics tend to be used.
Conclusions
This research profiled antibiotic resistance patterns of bacterial isolates obtained from live bird’s market in Sokoto, northwestern Nigeria, a part of the world with little data to address the effect of farming practices on antibiotic resistance. Of particular concern was that genera containing pathogenic members were the most commonly identified and that many of the antibiotic resistance patterns indicated multiple antibiotic resistance and it is a potential for antibiotic treatment failures. Penicillin G resistance was common among all isolates. Complete resistance to all the test antibiotics was observed in one of the Enterobacter spp., and this could be due to possession of resistant genes. It was also observed that almost all the isolates were highly susceptible to gentamicin. However, the free-range type of practice may increase the potential for transfer of resistant gene to humans. For example, free range farming practices, may present benefits from high turnover rates, but there is increased potential for human contact with birds. It could be concluded that the organisms may pass through the faeces of birds to the environment and cause potential human health hazards and can cause illnesses.
Recommendations
It is recommended that the resistant isolates should be save and sent to the reference laboratories for additional testing, this is because some organisms phenotypically may appear to be resistant while genetically they may not be resistant. It is also recommended that susceptible agents should be studied further clinically because some agents may appear susceptible in vitro but may not susceptible clinically (in vivo). Further study is recommended to demonstrate which farming practices best reduce antibiotic resistance levels and minimize the potential to spread to humans.
References
- Taylor LH, Latham SM, Woolhouse MEJ (2001) Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356(1411): 983-989.
- Belshe RB (1998) Influenza as a zoonosis: how likely is a pandemic?. Lancet 351(9101): 460-461.
- Reed KD, Meece JK, Henkel JS, Shukla SK (2003) Birds, migration and emerging zoonoses: West Nile virus, lyme disease, influenza and enteropathogens. Clinical Medicine & Research 1(1): 5-12.
- Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, et al (2007) The genome sequence of avian pathogenic Escherichia coli strain o1: K1: H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. Journal of Bacteriology 189(8): 3228-3236.
- Fournier P E, Tissot Dupont H, Gallais H, Raoult D (2000) Rickettsia mongolotimonae: A rare pathogen in France. Emerging Infectious Diseases 6(3): 290-292.
- Rappole JH, Derrickson SR, Hubálek Z (2000) Migratory birds and spread of West Nile virus in the western hemisphere. Emerging Infectious Diseases 6(4): 319-328.
- Humair P F (2002) Birds and borrelia. International Journal of Medical Microbiology 291(Suppl 33): 70-74.
- Winker K, McCracken KG, Gibson DD, Pruett CL, Meier R, Huettmann F, et al (2007) Movements of birds and avian influenza from Asia into Alaska. Emerging Infectious Diseases 13(4): 547-552.
- Benskin CM, Wilson K, Jones K, Hartley IR (2009) Bacterial pathogens in wild birds: a review of the frequency and effects of infection. Biological Reviews 84(3): 349-373.
- Altizer S, Bartel R, Han BA (2011) Animal migration and infectious disease risk. Science 331(6015): 296-302.
- Waters A, Higgins D, McCutchan T (1991) Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc Natl Acad Sci U S A 88(8): 3140-3144.
- Capua I, Alexander DJ (2002) Avian influenza and human health. Acta Tropica 83(1): 1-6.
- McKinney ML (2002) Urbanization, biodiversity, and conservation the impacts of urbanization on native species are poorly studied but educating a highly urbanized human population about these impacts can greatly improve species conservation in all ecosystems. BioScience 52(10): 883-890.
- Atterby C, Ramey AM, Hall GG, Järhult J, Börjesson S, et al (2016) Increased prevalence of antibiotic-resistant E coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infection Ecology & Epidemiology 6: 32334-32339.
- Tsiodras S, Kelesidis T, Kelesidis I, Bauchinger U, Falagas ME (2008) Human infections associated with wild birds. Journal of Infection 56(2): 83-98.
- Bonnedahl J, Drobni M, Gauthier Clerc M, Hernandez J, Granholm S, et al (2009) Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One 4(6): e5958.
- Literák I, Kulich P, Robesova B, Adamik P, Roubalova E (2010) Avipoxvirus in great tits (Parus major). European Journal of Wildlife Research 56: 529-534.
- World Health Organization (2013). Manual of Basic Techniques for Health Laboratory. 2nd ed. Geneva: World Health Organization.
- (1981) Council for Agricultural Science and Technology. Antibiotics in Animal Feed.
- (2000) Office International des Epizooties (OIE), Manual of standards for diagnostics tests and vaccines.
- Cheesbrough M (1985) Medical laboratory manual for tropical countries. I.stedi. Vol 2. Microbiology. English Language Book Society, London pp. 400-480.
- (2009) Clinical and laboratory standard institute (CLSI), Performance standard for antimicrobial susceptibility test.
- Adeleke E, B Omafuvbe (2011) Antibiotic resistance of aerobic mesophilic bacteria isolated from poultry faeces. Res J Microbiol 6(4): 356-365.
- Pei R, S C Kim, K H Carlson, A Pruden (2006) Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40(12): 2427-2435.
- Van den Bogaard A, London N, C Driessen, E Stobberingh (2001) Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J Antimicrob Chemother 47(6): 763-771.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...