Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-5945
Research Article(ISSN: 2638-5945) 
Studies on Hybridization and Genetic Diversity in Cowpea (Vigna unguiculata L) Volume 2 - Issue 2
T Rangkham and VK Khanna*
- School of Crop Improvement, College of Post-Graduate Studies, Central Agricultural University, Meghalaya, India
Received: June 12, 2018; Published: June 18, 2018
Corresponding author: VK Khanna, School of Crop Improvement, College of Post-Graduate Studies, Central Agricultural University, Meghalaya, India
DOI: 10.32474/OAJOM.2018.02.000132
Abstract
Crossability of various accessions of cowpea on the basis of pollen development and fruit set was studied along with genetic diversity by SSR markers. There was a significant correlation between fruit set and percent viable pollen. In all selfings and the crosses, pollen germination increased from 2 hr to 6 hr after pollination. Pollen germination had a highly significant correlation with fruit set. On selfing, the fruit set was positively correlated with faster pollen tube growth whereas it was not so in the case of crosses. There was a positive correlation between pollen tube growth and fruit set. Thirty six primers were selected to assess genetic diversity of 36 accessions of cowpea and twenty seven primers were found to be polymorphic. The polymorphic primers generated a total of 72 alleles. The number of alleles ranged from 2 to 4 with an average of 2.66 per locus. Based on Euclidean similarity matrix and dendrogram constructed, two major clusters were revealed. Cluster I and Cluster II comprised of 14 and 22 accessions, respectively. M-5 was found to be more distinct from Kashi Gowri, EC- 559390 and EC- 9738; EC- 472260 and Sel- 16; EC- 30950 and PL- 2; EC- 472260 and EC- 9739. Sel- 16 and EC- 9739 were found to be very less distinct. The value of Euclidean similarity coefficient ranged from 0 to 3.61. PIC value obtained ranged from 0.09 – 0.68 with an average of 0.35. Based on PIC values, out of 27 primers, SSR 6314 was found to be highly informative.
Keywords: Crossability, Vigna unguiculata, Pollen Tube, Fruit Set, SSR
Introduction
Cowpea (Vigna unguiculata L.) is a self-pollinated crop belonging to the family Fabaceae and is native to Central Africa. It is also known as the ‘poor man’s meat’ because of its good protein quality and high nutritional value. Legumes are economically significant crops and rich source of nourishing diet for growing population [1,2]. Reported that the floral bud initiation and development is sensitive to photoperiod in many cowpea accessions, and in some genotypes the degree of photoperiod sensitivity (i.e. the extent of delay in flowering) is influenced by temperature [3]. Also reported that the productivity of cowpea (Vigna unguiculata) can be reduced substantially by hot weather during flowering.
The assessment of pollen viability is important in artificial pollination and breeding experiments [4,5] stated that for fertilization to occur, the pollen landing on the stigma must be viable i.e. which must be able to germinate, penetrate the stigma and style and reach the ovule during the effective pollination period (EPP). For successful fertilization to occur there should be components of pollen performance which includes pollen produced in a flower, pollen morphological homogeneity, pollen germination, pollen tube growth and pollen competition [6-8] stated that the pollen tube growth and pollen germination are important research materials for morphological, physiological, biotechnological, ecological, evolutional, and molecular studies. Intraspecific hybrids are an important source of variability in breeding programs. The knowledge of the intraspecific compatibility as well as of the direction of crossing is extremely important for success in hybrid breeding programs [9]. Genetic diversity analysis facilitates utilization of genetic resources for conservation of germplasm and the development of breeding programs [10]. Cowpea has a very narrow gene pool which limits development of new cowpea varieties by cross-breeding [11]. The knowledge of genetic diversity of cowpea germplasm is extremely essential for cowpea breeders to produce new cowpea varieties with higher yield and better quality [12]. This paper reports pollen germination and pollen tube behavior in relation to crossability between various accessions of Vigna unguiculata L. and seed set and genetic diversity using SSR primers.
Materials and Methods
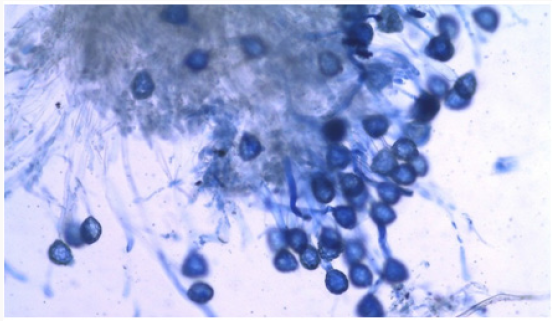
The 36 accessions used were Kashi Gowri, Kashi Kanchan, IC- 202826, EC- 30950, PL- 2, PL- 3, Kashi Nidhi, IC- 33922, Arka Garima, Kashi Unnati, PL- 4, PL- 1, PL- 5, EC- 472260, Sel- 16, IC- 559390, EC- 9738, IC- 3004, IC- 202865, EC- 9739, EC- 110598, Kashi Shyawal, IC- 332198, EC- 390221, EC- 15296, EC- 9736 and EC- 390211 and accessions of N- 1, PS- 1, CP- 7, HS, M- 1, M- 2, M- 3, M- 4 and M- 5 were collected from different locations of North Eastern region of India. Five parents were selected for crossability studies. They were used for one way crossing and so the total number of selfing and crossing were 15. For each cross at least ten to fifteen flowers were randomly selected from plants. Pollinations were performed from May to October, 2016 and care was taken to avoid any damage to the stigma. The styles were collected 2 to 6 hr after hand pollination and fixed immediately in 1:3 glacial acetic acid-ethyl alcohol for at least 24 hr and then preserved in 70 per cent alcohol till further use. For pollen germination and pollen tube growth observations, the pollinated flowers were gently rinsed in distilled water and pistils were separated from the flowers after which they were kept in a drop of 1N HCl for 10 minutes. They were again rinsed in distilled water and stained in 1 percent aniline blue [13,14]. The time required for staining was 10-20 seconds depending on the thickness of the style and the stage of penetration of the pollen tube in the stigma. After staining, the pistils were destained for 20-24 hr in a 1:1:1 mixture of 40 % acetic acid: orthophosphoric acid: distilled water. The pistils were then rinsed in distilled water and mounted in pure lactic acid and studied under the microscope. The pollen grains and pollen tubes stained deep blue.
Genomic DNA from cowpea leaves was isolated using CTAB method of [15]. Young, actively growing leaves of 5-10 days old plants were collected and used for DNA extraction. The quantification of DNA were done by staining DNA with ethidium bromide after electrophoresis in 0.8% agarose gel at 80 V for 1 hr in TBE buffer (0.04 M Tris borate, 0.001 M EDTA, pH 8.0) using known DNA concentration standards. Molecular weights of bands were estimated by using 100 bp for SSR. The homology of bands was based on the distance of migration in the gel. SSR amplicons obtained from each entry were resolved as multiple and a single band, respectively on the agarose system and the pair-wise correlation matrix was developed on the normalized data set to find out the relationship among 36 accessions of cowpea. Bands were marked: presence of a band was scored as one (1) and absence of a band as zero (0). Very faint bands were not considered for final scoring. After scoring, the Polymorphism Information Content (PIC) was calculated. The SSR data was subjected to Paleontological Statistics, PAST v3.15 software [16] to construct a Dendrogram by hierarchical cluster analysis based on Ward’s method (Ward, 1963) using the Euclidean similarity matrix [17] as cited by [18].
Results and Discussion
Pollen Fertility
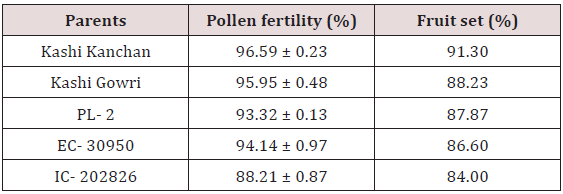
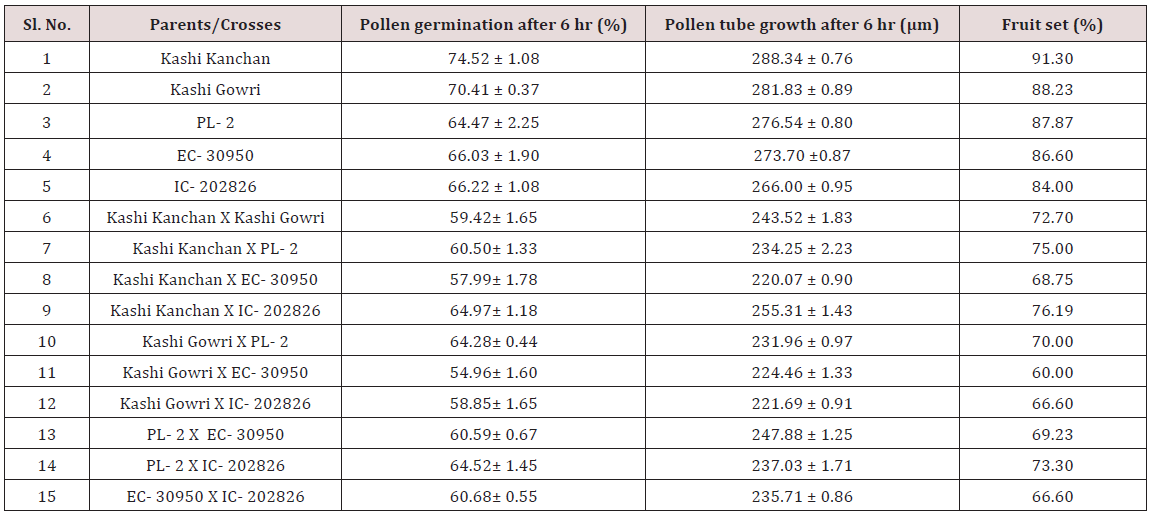
The pollen fertility of all the accessions under this study revealed that the maximum fertile, stained pollen was recorded in Kashi Kanchan (96.59 ± 0.23) whereas the lowest percentage of fertile pollen recorded was in IC- 202836 (88.21 ± 0.87) (Table 1). However, the average percent pollen fertility did not show too much difference among the accessions [19] stated that in cowpea, high night temperature (33/30º C) increased the occurrence of small and shrunken pollen leading to zero pod set compared with low night temperature (33/20º C) (Figures 1-3).
Pollen Germination
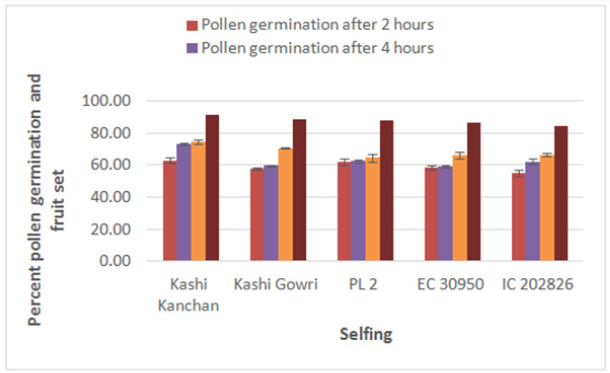
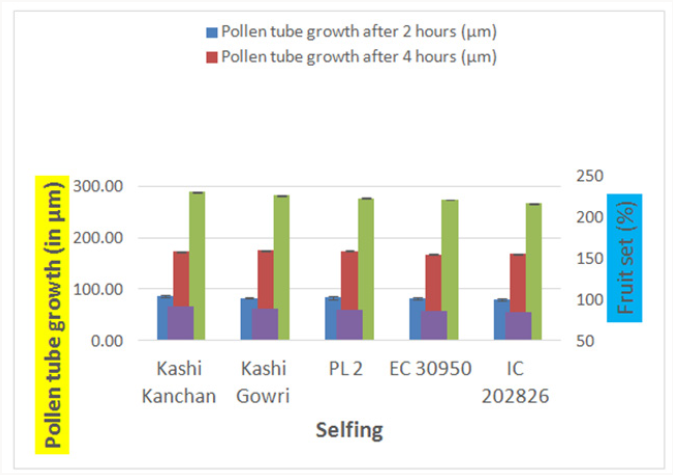
Maximum pollen germination after 2 hr of pollination was observed in selfing of Kashi Kanchan (62.93 %) and the least pollen germination was recorded in cross of Kashi Kanchan and EC- 30950 (49.47 %). The results show that there was a constant increase in pollen germination from 2 hr to 6 hr in all the parents. After 2 hr of pollination, on selfing, maximum pollen germination was recorded in Kashi Kanchan (62.93 %) followed by PL- 2 (61.97 %), EC- 30950 (58.31 %), Kashi Gowri (57.98 %), and IC- 202826 (55.21 %). After 4 hr of pollination on selfing, maximum pollen germination was recorded in Kashi Kanchan (73.18 %) and the least was recorded in EC- 30950 (59.06%). After 6 hr of pollination on selfing, maximum pollen germination was recorded in Kashi Kanchan (74.52 %) and the least was recorded in PL- 2 (64.47 %).
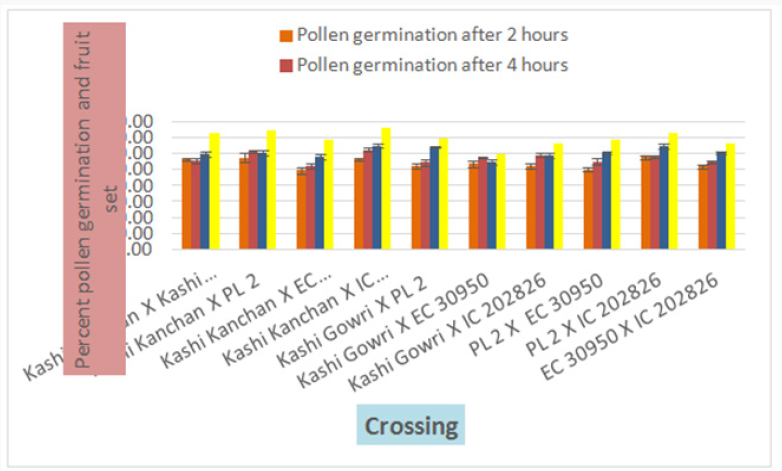
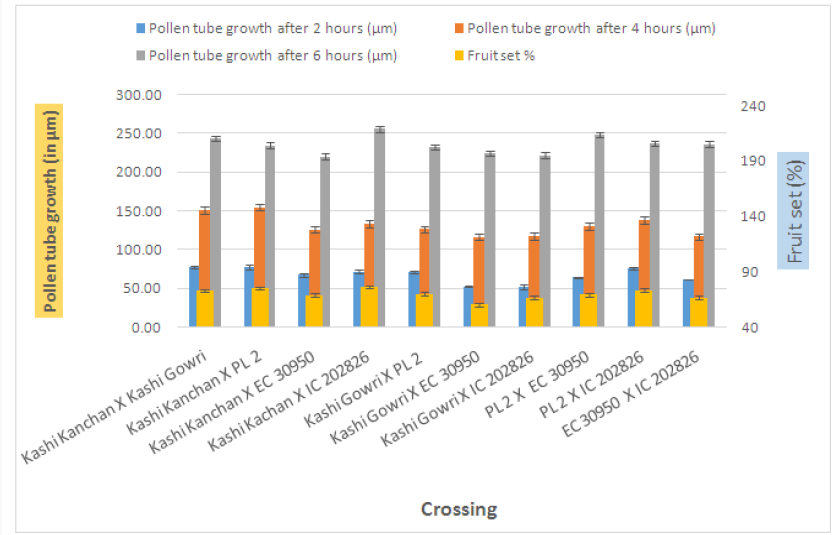
In crosses between the parents, the maximum pollen germination was recorded in PL- 2 X IC- 202826 (57.88 %) after 2 hr of pollination and the least was found in Kashi Kanchan X EC- 30950 (49.47 %). After 4 hr of pollination, maximum pollen germination was recorded in Kashi Kanchan X IC- 202826 (62.40 %). The least germination was recorded in Kashi Kanchan X EC- 30950 (52.40%). After 6 hr of pollination, maximum pollen germination was recorded in Kashi Kanchan X IC- 202826 (64.97%) and the least was recorded in Kashi Gowri X EC- 30950 (54.96%). It was observed that pollen germination had significant correlation with fruit set. Similar results have been reported by [20,21] in wild accessions of chillies and tomato, respectively.
Pollen Tube Growth
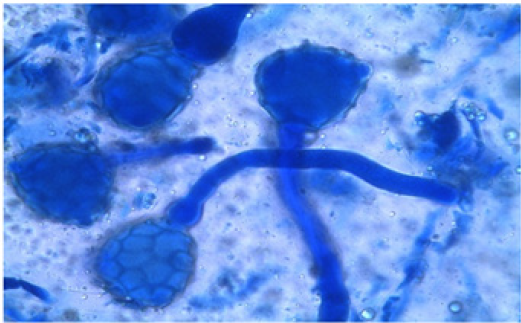
The emergence of pollen tubes on the stigma and the style was seen after different time intervals on selfing and in crosses. Pollen fall on the stigma of each individual flower was variable because the amount of pollen grains differ during the hand pollination and it also depends on how many will stick on the stigma. The length of pollen tubes was measured (μm) at three different time intervals on selfing as well as in crosses. Length of pollen tube was recorded after staining with 1 % aniline blue stain. At 2 hr after pollination, a number of pollen grains had germinated and the tubes penetrated into the stigma hairs. Some of the pollen tubes had just started their growth in the hairs and a few had already entered the transmitting tissue of the style. After 2 hr of selfing the maximum pollen tube growth was observed in Kashi Kanchan (86.90 μm) and the minimum in IC- 202826 (80.19 μm). Pollen tubes can act cooperatively during growth in the style to increase fertilization success [22] and they can also compete with each other to fertilize the available egg cells [23].
In case of crosses, the maximum pollen tube growth was observed in Kashi Kanchan X PL- 2 (77.11 μm) and the minimum was observed in Kashi Gowri X IC- 202826 (51.16 μm). After 4 hr of pollination, there was a greater differentiation and elongation in the pollen tubes. Some had just started to elongate and all intermediate stages were observed. On selfing, maximum pollen tube growth was observed in Kashi Gowri (174.91 μm) and the least in EC- 30950 (167.53 μm) at 4 hr after pollination. In crosses, maximum pollen tube growth was observed in Kashi Kanchan X PL- 2 (154.32 μm) as at 2 hr after pollination and the least in Kashi Gowri (116.16 μm). On selfing, maximum pollen tube growth after 6 hr of pollination was observed in Kashi Kanchan (288.34 μm) and the minimum was observed in EC- 30950 (273.70 μm). In crosses, maximum pollen tube growth was observed in Kashi Kanchan X Kashi Gowri (243.52 μm) and minimum pollen tube growth was observed in Kashi Kanchan X EC- 30950 (220.07 μm). So, the results were quite similar at 2 hr, 4 hr and 6 hr after pollination (Figures 4-6).
Fruit Set
The maximum fruit set was obtained in selfing as compared to crosses. Among the selfing, the maximum fruit set was obtained in Kashi Kanchan (91.3 %) and the least in IC- 202826 (84 %). When the parents were crossed, the maximum fruit set was obtained in Kashi Kanchan X IC- 202826 (76.19 %) and the least fruit set was obtained in Kashi Gowri X EC- 30950 (60 %). In general, fruit set was less in crosses. The results indicated that cowpea flowers are self-fertile, and crosses in cowpea make good cross compatability, which makes the successful formation of fruit set. This is similar to the results of Elmobarak (2005). This also reveals that crossing between the accessions was successful, as normal pod set was observed [24]. Reported that out crossing in flowering plants avoids the deleterious effects of inbreeding depression and promotes heterozygosity, genetic variability, and genetic exchange, which in turn leads to the long-term survival and adaptation of a species (Table 2) (Figures 7-10).
Correlation Studies in Crosses of Vigna unguiculata L
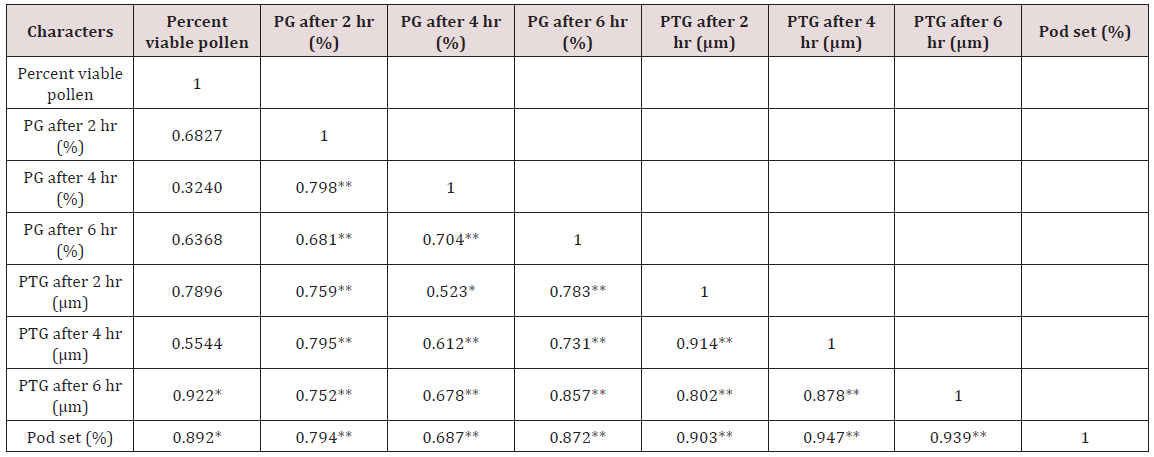
Correlation studies on pollen germination recorded nonsignificant values for percent viable pollen. However, pollen germination after 4 hr of pollination was highly correlated with pollen germination after 2 hr of pollination and pollen germination after 6 hr of pollination had a highly significant correlation with pollen germination after 2 hr and 4 hr of pollination, respectively. Pollen tube growth after 6 hr of pollination recorded significant correlation with percent viable pollen. Pollen tube growth after 2 hr of pollination was observed to be significantly correlated with pollen germination after 4 hr of pollination and highly correlated with pollen germination after 2 hr and 6 hr of pollination. Pollen tube growth after 4 hr of pollination was highly significant with pollen germination after 2 hr, 4 hr and 6 hr of pollination, and also with pollen tube growth after 2 hr of pollination. Pollen tube growth after 6 hr of pollination correlated with percent pollen fertility at 5 % level of significance and highly correlated with pollen germination after 2 hr, 4 hr and 6 hr of pollination and also with pollen tube growth after 2 hr and 4 hr of pollination. Fruit set had highly significant correlation with pollen germination (0.794, 0.687 and 0.872). A positive correlation with pollen tube growth was seen which was highly significant at 1 % level of significance (0.903, 0.947 and 0.939). At 5 % level of significance, positive correlation was seen between fruit set and percent viable pollen (0.892) (Table 3).
PG= pollen germination; PTG= pollen tube growth.
Note: *denotes significant at 5 % level of significance, ** denotes significant at 1 % level of significance.
SSR Analysis
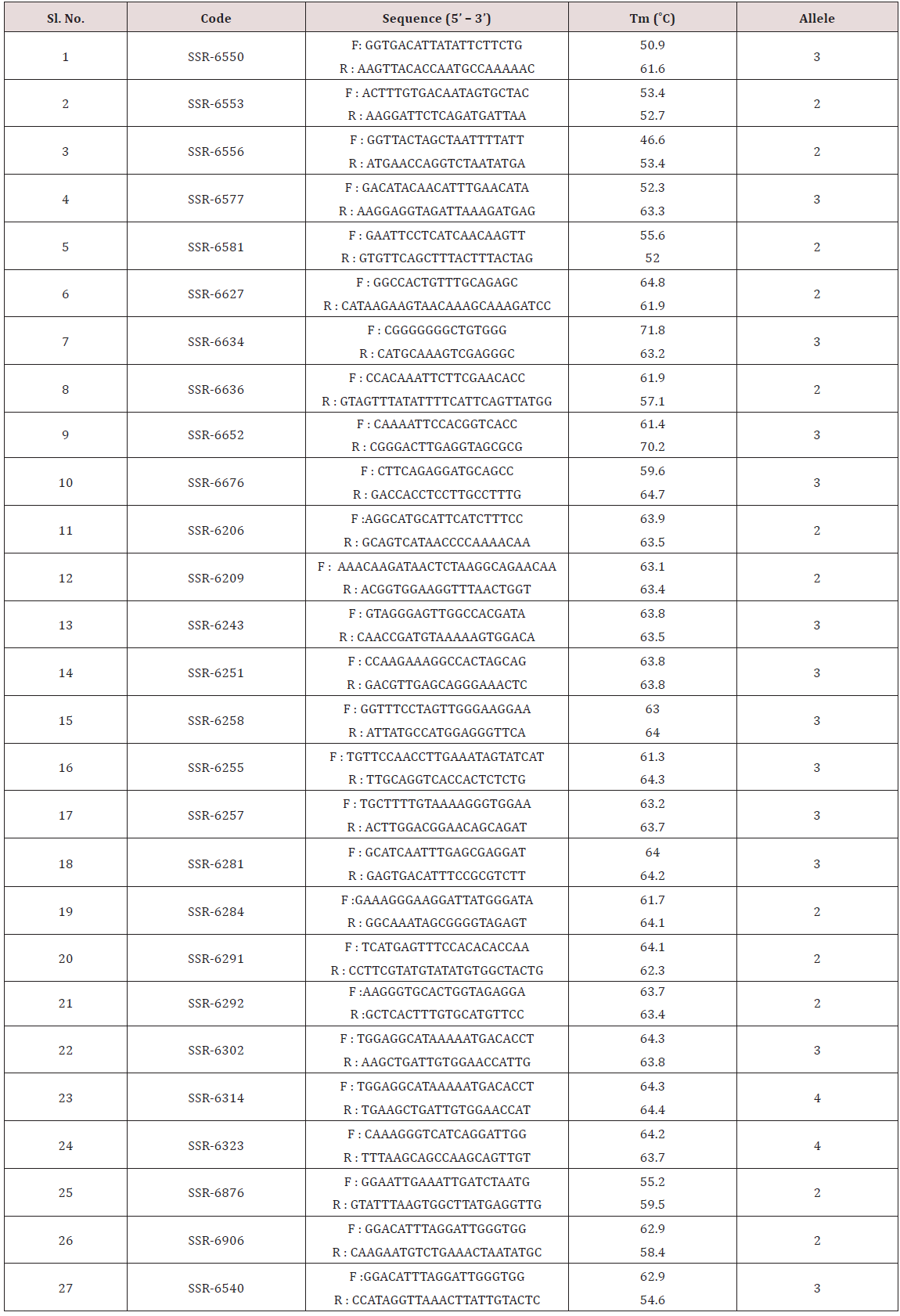
Thirty six primers were used for genetic diversity analysis and twenty seven primers were found to be polymorphic. The polymorphic primers generated a total of 72 alleles. The data obtained from SSR analysis were subjected to Euclidean similarity analysis to find out the relationship among the accessions analyzed. The value of Euclidean similarity coefficient ranged from 0 to 3.61. Based on Euclidean similarity matrix it was found that the lowest similarity coefficient was found between M-5 and Kashi Gowri (3.61) whereas EC- 559390 and EC- 9738; EC- 472260 and Sel- 16; EC- 30950 and PL- 2; EC- 472260 and EC- 9739; and Sel- 16 and EC- 9739 showed the highest similarity coefficient (0) (Table 4).
From the dendrogram generated using Paleontological Statistics, PAST v3.15 software, the genotypes were grouped into two major clusters. The first major cluster was divided into I A and I B sub- clusters. The second major cluster II was divided into II A and II B sub- clusters. The sub- cluster I A consisted of EC-559390 and EC-9738 which are highly similar to each other. Sub- cluster I B was divided into two small sub- clusters I B I and I B II. Five accessions were grouped into sub-cluster I B I and Kashi Kanchan appears to be the most distinct from all the other in sub- cluster I B I. I B II sub- cluster comprise of 7 accessions and EC 472260 and Sel-16 are highly similar to each other. Cluster II was divided into two II A and II B sub- clusters. II A comprises of four accessions and Kashi Gowri appears to be most distinct from all the others in sub- cluster II A and EC- 30950 and PL- 2 are highly similar to each other. Cluster II B again was further divided into II B I and II B II sub- clusters. Sub- cluster II B I comprises of 5 accessions and M-1 appears to be the most distinct from all the others in sub- cluster II B I. Thirteen accessions are grouped in small sub- cluster II B II and in this accessions, Kashi Shyawal and EC-390221 appear to be more distinct.
For an effective breeding programme, information concerning the extent and nature of genetic diversity within a crop species is essential [25]. Accurate assessment of genetic variability is important for the preservation and utilization of germplasm resources (Huaqiang et al., 2012). The knowledge of the genetic diversity available within the local and regional germplasm collections can enhance the overall effectiveness of cowpea improve¬ment programs (Hegde and Mishra, 2009). The use of genetic diversity in breeding program is important to decrease crop vulnerability to abiotic and biotic stresses [26].
Figure 11: (A) and (B). SSR profiles generated by SSR 6257 and SSR 6314 primers, respectively on 36 accessions of Vigna unguiculata. Number 1-36 refers to genotypes, M indicates 100 bp ladder.
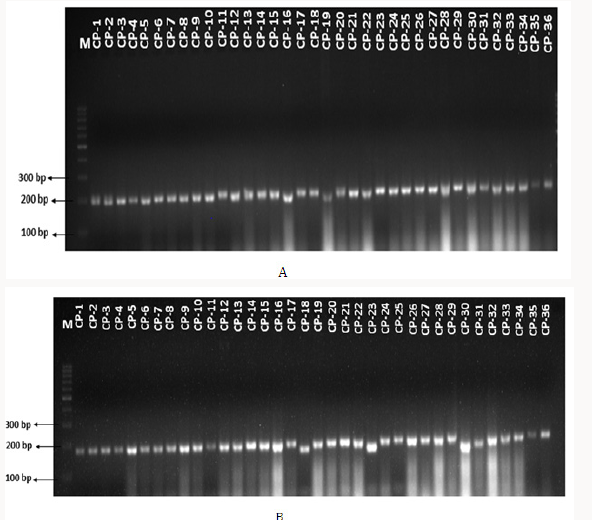
Figure 12: Dendrogram generated using Ward’s method showing relationship between 36 accessions of cowpea.
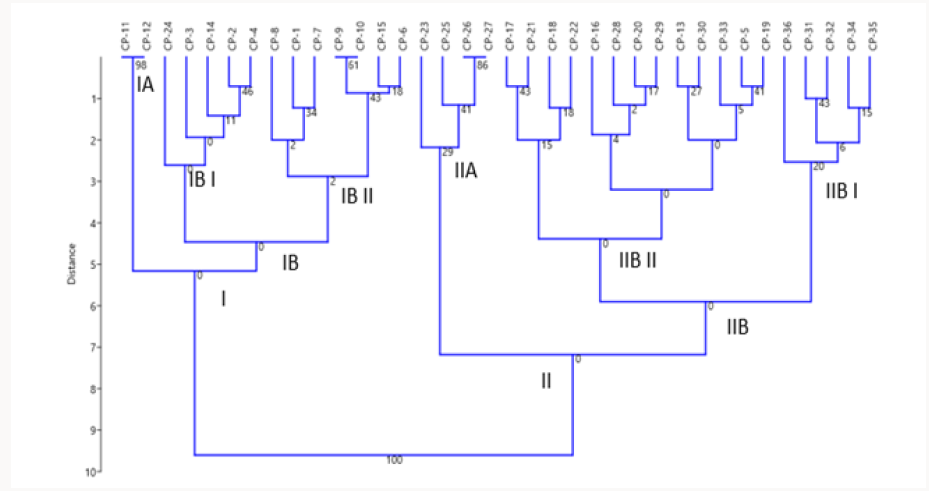
In the present study genetic diversity among 36 accessions of cowpea was assessed by using 36 SSR markers (Figures 11 A & 11B). A total of 72 alleles were detected using 27 polymorphic SSR loci with the mean value of 2.66 per locus. The alleles number was lesser than that reported by [27] who detected 249 alleles with the mean value of 5.08 per locus. The polymorphism information content varied from 0.09 to 0.68 with an average of 0.35. This was similar to that reported by [28] who analyzed genetic diversity in 141 accessions of cowpea using 25 SSR markers. They found PIC value ranging from 0.07 to 0.66 with an average of 0.38. Based on PIC values, out of 27 primers, SSR 6314 was found to be highly informative. When compared with the study of [27] the allele’s number was lesser which may be due to the use of more number of primers and the PIC value found in the study was more than the PIC value found by [27]. The Euclidean similarity coefficient for all 36 accessions ranged from 0 to 3.61. However, some of the accessions showed similarity value of 0. This may be due to lesser number of SSR primers used in the present study [29-32]. Cluster analysis performed based on Euclidean similarity matrix generated two major clusters. Cluster I and cluster II comprised of 14 and 22 accessions, respectively. Cluster I was divided into I A and I B sub clusters. The sub cluster II B was further divided into two small clusters .Cluster II was also divided into II A and II B sub clusters. The sub cluster II B was further divided into two small clusters (Figure 12).
References
- Wein HC, Summerfield RJ (1980) Adaptation of cowpeas in West Africa: Effects of photoperiod and temperature responses in cultivars of diverse origin. In: Summerfield RJ Bunting, AH (Eds.), Advances in Legume Science. Royal Botanic Gardens, Kew, UK, pp. 405-417.
- Ehlers JD, Hall AE (1996) Genotypic classification of cowpea based on responses to heat and photoperiod. Crop Sci 36: 673-679.
- Marfo KO, Hall AE (1992) Inheritance of Heat Tolerance during Pod Set in Cowpea.
- Rodriguez-Riano T, Dafni A (2000) A new procedure to assess pollen viability. Sex Plant Reprod 12(4): 241-244.
- Shivanna KR, Linskens HF, Cresti M (1991) Responses of tobacco pollen to high humidity and heat stress: viability and germinability in vitro and in vivo. Sex Plant Reprod 4: 104-109.
- Thompson M (2004) Flowering, pollination and fruit set. In: Webster AD, Looney NE (Eds.), Cherries, Crop Physiology, Production and Uses. Wallingford, CABI Publishing, pp. 223-243.
- Janick J and Moore NJ (1996) Fruit Breeding, Tree and Tropical Fruits. John Wiley and Sons, New York, USA.
- Dane F, Olgun G, Dalgic O (2004) In vitro pollen germination of some plant species in basic culture medium. J of Cell andMolecular Biology 3: 71-76.
- Nascimento NFF, Nascimento MF, Rego ER, Lima JAM, Rego MM, et al. (2015) Intraspecific cross-compatibility in ornamental pepper.
- Hu J, Wang P, Su Y, Wang R, Li Q, et al. (2015) Microsatellite diversity, population structure, and core collection formation in melon germplasm. Plant Mol Biol Rep 33: 439-447.
- Staub JE, Chung SM, Fazio G (2005) Conformity and genetic relatedness estimation in crop species having a narrow genetic base: the case of cowpea (Vigna unguiculata LL) Plant Breeding 124: 44-53.
- Innark P, Khanobdee C, Samipak S, Jantasuriyarat C (2013) Evaluation of genetic diversity in cowpea (Vigna unguiculata L. L.) germplasm using agro-economic traits and microsatellite markers. Sci. Hortic 162: 278- 284.
- Gerlach D (1969) A rapid safranine-crystal violet-light green staining sequence for paraffin sections of plant materials. Stain Tech 44(4): 210- 211.
- D’Souza L (1972) Staining pollen tubes in styles of cereal with cotton blue: Fixation by ethanol-lactic acid for enhanced differentiation. Stain Tech 47: 107-108
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissues. Focus 12: 13-15.
- Hammer O, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics software package for education and data analysis. Palaeontologia Electronica 4: 9.
- Cruz CD, Regazzi AJ (1994) Modelos Biométricos Aplicados ao Melhoramento Genético. Imprensa Universitária, Viçosa, pp. 390.
- Oliveira MAP, Valls JFM (2003) Morphological characterization and reproductive aspects in genetic variability studies of forage Peanut, Scientia Agricola 60(2): 299-304.
- Ahmed FE, Hall AE, DeMason DA (1992) Heat injury during floral development in cowpea (Vigna unguiculata). American Journal of Botany 79(7): 784-791.
- Debbarama C, Khanna VK, Tyagi W, Rai M, Meetei NT (2013) Wide Hybridization and Embryo-Rescue for Crop Improvement in Capsicum. Agrotechnol.
- Kharkongar HP, Khanna VK, Tyagi W, Rai M, Meetei NT (2013) Wide Hybridization and Embryo-Rescue for Crop Improvement in Solanum. Agrotechnol.
- Esser K (1953) Z Indukt Abstain Vererb 85: 28-50.
- Mulcahy DL, Mulcahy GB (1975) The influence of gametophytic competition on sporophytic quality in Dianthus chinensis. Theor Appl Genet 46: 277-280.
- Dellaporta SL, Urrea AC (1993) Sex Determination in Flowering Plants. The Plant Cell 5: 1241-1251.
- Demir K, Bakir M, Sarikamis G, Acunpalp S (2010) Genetic diversity of egg plant (Solanum melongena) germplasm from Turkey assessed by SSR and RAPD markers. Genetics and Molecular Research 9(3): 1568- 1576.
- Doumbia IZ, Akromah R, Asibu JY (2014) Assessment of Cowpea Germplasms from Ghana and Mali Using Simple Sequence Repeat (SSR) Markers. International Journal of Agriculture and Forestry, 4(2): 118- 123.
- Badiane FA, Gowda BS, Cisse N, Diouf D, Sadio O, et al. (2012) Genetic relationship of cowpea (Vigna unguiculata) varieties from Senegal based on SSR markers. Genet Mol Res 11(1): 292-304.
- Asare AT, Gowda BS, Galyuon IKA, Aboagye LL, Takrama JF, et al. (2010) Assessment of the genetic diversity in cowpea (Vigna unguiculata L Walp) germplasm from Ghana using simple sequence repeat markers. Plant Genetic Resources: Characterization and Utilization, pp. 1- 9.
- Elmobarak EAE (2005) Studies on Floral Biology in Snake melon. MSc Thesis, Submitted to Omdurman Islamic University.
- Hegde VS, Mishra SK (2009) Landraces of cowpea, Vigna unguiculata (L.) Walp., as potential sources of genes for unique characters in breeding. Genet Res Crop Evol 56(5): 615-627.
- Huaqiang T, Manman T, Qian L, Yongpeng Z, Jia L, et al. (2012) A Review of Molecular Markers Applied in Cowpea (Vigna unguiculata L Walp). Breeding J Life Sci 6: 1190-1199.
- Ward Jr JH (1963) Hierarchical grouping to optimize an objective function. Journal of the American statistical association 58(301): 236- 244.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...