Lupine Publishers Group
Lupine Publishers
Menu
Review Article(ISSN: 2641-6875) 
Shift of The Neutral-Niche Dynamics in The Human Microbiome Under Disturbances Associated with Lifestyle Changes Volume 1 - Issue 5
Huang Qi1, Yinlei Liao1 and Yao Xia2*
- 1The First Affiliated Hospital of Kunming Medical University, Kunming, China
- 2Kunming Institute of Zoology, Kunming, China
Received: March 14, 2020; Published: June 19, 2020
*Corresponding author: Yao Xia, Kunming Institute of Zoology, Kunming, China
DOI: 10.32474/CTBM.2020.01.000125
Abstract
Background: It is well recognized by now that human micro biome has far reaching influences on our healthy and diseases. One venue to investigate the influences can be through the study of the mechanisms of microbial community assembly and diversity maintenance. In this article, we apply a hybrid model of neutral-niche dynamics by Ofiteru et al. to reanalyze the dataset from David et al.’s study on the effects of life style to the micro biome of two individuals, aiming to explore the shift of the neutral-niche dynamics in the human micro biota.
Results: We obtained two major findings. First, in gut, Faecali bacterium prausnitzii, an anti-inflammatory bacterium, exhibited high neutral dynamics, and few other species showed similar level of neutral dynamics. In contrast, in the oral micro biome, more species demonstrated high neutral dynamics. Second, we detected some bacterial species whose neutral-niche dynamics are strongly associated with the occurrences of disturbances such as change of diets and infection.
Conclusions: We postulate that the difference between gut and oral micro biota could be due to the open gateway nature of oral habitat, which makes it difficult for niche-selected species to maintain. The balance shift in the neutral-niche dynamics may play an important role in dysbiosis and possess broad health implication.
Background
The great significance of human microbiota on our health and
diseases been recognized increasingly thanks to the human micro
biome projects [1,2]. For examples, microbiota offer critical services
such as protecting against diseases [3,4] stimulation of intestinal
angiogenesis [5] and regulation of host fat storage [6], which are
critical for our health. In the mean time, micro biota is thought to
relate to a number of diseases, such as inflammatory bowel disease
[7,8], colon cancer [8,9], obesity Le et al. [10] and type-2 diabetes
Qin et al. [11]. Though human microbiota are relatively stable in
the long-term scale [12,13], factors such as use of antibiotics
[14-16], unhealthy diets [17,18] and diseases [19,20] and other
disturbances are likely to break the balance and may cause the sotermed
dysbiosis-state of micro biome associated with diseases.
Theoretically, the mechanism of community assembly and
diversity maintenance should have far reaching influence on the
services of human micro biome on its host, but our understanding
of the mechanism per se has been very limited. Traditionally, there
have been two diametrically opposite opinions on this issue, i.e.,
deterministic niche theory and stochastic neutral theory [21-
26] and extensive investigations have been conducted, including
reconciling both theories in various frameworks of hybrid models
in macro ecology of plants and animals. The forces that govern the
formation of micro biota is supposed to be controlled by a number
of deterministic factors, such as host species, genotype, diet, health
and interactions between microbes [27], which seem to support
the niche theory. The fundamental assumption of niche theory is that the ecological traits vary among different species [28]. The
alternative neutral theory that is proposed by Hubbell [23] and
has been applied in ecology area for years brings a novel idea
that micro biota assembly may be driven by stochastic process of
dispersal and ecological drift, of which the assumption is that all
species are ecologically equivalent (i.e. all species have equal birth,
death, dispersal and speciation rates) [29].
In this study, we apply one of the hybrid frameworks originally
developed by Ofiţeru et al. [31] to further investigation the possibly
combined niche and neutral effects in the human gut micro biota.
Ofiteru et al. [31] argued that neutral dynamics should play an
important role in the community assembly if the same neutral
models could explain the dynamics of populations in a community.
Specifically, they recalibrated and validated a purely neutral model
and an extended parsimonious model including environmental
factors to fit time-series of populations in a microbial wastewater
treatment community. Another factor motivated Ofiteru et al.
(2010) ‘recalibration’ is their critic to the most common method
for testing the neutral theory. They argue that (i) the commonly
used methods are not robust enough because different parameters
pairs could result in similar species abundance distribution (SAD);
(ii) the relative importance of niche and neutral forces are difficult
to differentiate, since the niche process and neutral process are
probably jointly responsible for the community assembly Dumbrell
et al. [32], Stegen et al. [33]; (iii) neutral models may fail to identify
the effects of environmental factors when neutral process exert
partly.
The dataset Ofiteru et al. [31] originally used to validate their
calibrated model was from Wells et al.’s [34] study on the micro
biota in a carefully managed wastewater treatment plant with
well-controlled environment, which appears to be a relatively
stable habitat for microbes and the neutral dynamics model is
likely to perform better. However, it has not be tested that in a
relatively unstable ecosystem such as the human gut where sudden
disturbances such as diseases or change of lifestyle may occur
relatively frequently. Therefore, we set two objectives in this study.
The first objective is to investigate the neutral-niche dynamics for
the most common species in the micro biota of human body that is a
less stable environment where niche effect could be more strongly
dominant. Second, we would like to explore the environmentdriven
alterations of neutral-niche dynamics for the most common
species. To implement our study, we choose to reanalyze the
dataset from David et al. [35] study aiming at exploring the effects
of disturbance and lifestyle on the dynamics of human-associated
micro biota, in which they recruited two individuals and followed
and sampled them daily for a period of about one year. The
study was rather comprehensive and offered an ideal dataset for
conducting our investigation. Their original analysis was focused
on whole community level analysis and did not involve any analysis
of neutral dynamics at either community or population (individual
taxon) level.
Environmental effects and corresponding niche-neutral
dynamics affecting individual species may vary according to the
specific attributions of species. It is still possible that neutral
dynamics, at least partly, influence the time-series of populations
when observed samples failed to pass the neutrality test using
traditional methods. Hence, under our first objective, with Ofiteru
et al.’s method [31], we may identify the portion of variance of
the time-series of taxon populations driven by neutral dynamics.
Although the gut may be a relatively stable environment in our
daily life, both of the two individuals recruited in David et al. study
[35] had experienced dramatic environment disturbance during
the research period. One individual had traveled from a major
American metropolitan area to the capital of a developing nation in
Southeast Asia between days 71 and 122, exposed to new diet and
environment and got diarrhea twice during the traveling. Another
individual suffered from an infection caused by food poisoning
between days 151 to 159. By applying Ofiteru et al. [31] method,
we hope to explore whether the neutral dynamics would change
during or after the dramatic disturbances.
Methods
Dataset description and preprocessing
The dataset we reanalyzed was originated from David et al.
[35] study. In their study, 2 individuals were recruited, individual A
and B. For individual A, stool samples were collected daily between
days 0 and 346, named A-Gut group, and saliva samples were
collected daily between days 26 and 364, named A-Saliva group. For
individual B, stool samples were collected daily between days 0 and
252, named B-Gut group. For each sample, the V4 regions of 16S
rRNA genes were amplified and sequenced with Illumina GAIIx. The
time-series OTU tables of 3 groups were picked at 97% similarity
via QIIME analysis pipeline (v1.3). The data analysis, including the
calculation of OTU tables should refer to David et al. [35].
To make our analysis more informative biologically, we excluded
the species without confirmed names. The Ofiteru et al. [31]
model adopts a weighted least-squares regression to estimate the
parameters, where the weight of independent variable is [X(1-X)]-1
(X represents the independent variable), requiring the independent
variable (i.e., abundance of each OTU at every time point) to be a
non-zero value. To adapt to such requirement, we filtered out OTUs
with more than 5% zero occurrences in the time-series and for OUT
containing zero values (less than 5% zero occurrences), we added 1
to the abundance of time-series at all time potions.
Computational procedures of the neutral and hybrid models
We used a calibrated neural model by Ofiţeru et al. [31], derived from Hubbell neutral model and extended for time-series data of OTUs in the micro biota, where environmental effects can be added when necessary. The model assumes that the species in a local community originated from a source community (meta community), where the species abundance distribution (SAD) obeys a log-series distribution with a single shape-deciding parameter θ. The difference between SAD in local community and that in metacommunity could be described with a function of a pair of parameters, NT and m, where NT is the number of individuals in the hypothetic neutral local community and m is the probability that an individual in the local community would be replaced by an individual from the meta community rather than from local reproduction when it died. An advantage factor (α’) representing the effect of external environment factors on birth rate is added, therefore the neutral assumption would be broken if α’ was not 0. In other words, the model would represent a pure neutral dynamic when α’ was 0; when α’ was not 0, the model would be hybrid dynamics as it combined neutral dynamic and environmental effects. A stochastic differential equation (SDE) could be used to describe the dynamics of observed relative abundance of the i-th specie at time t, X(t), which is controlled by the parameters NTm and p, the relative abundance of the specie in the metacommunity. That is,
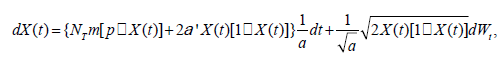 (1)
(1)where Wt is a Wiener process (standard Brownian motion) and a is an unknown constant that is related to the time between birth and death. For each of the dataset we tested, X(t) is known at successive 365 time points (one year) and dX(t) could be calculated as the longitudinal change in the relative abundance of a certain specie during the period. Eq. (1) could be approximated with a linear model, i.e.,
dX = m0 +m1Y1 +m2Y2 +e
where
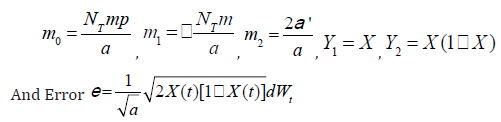 (1)
(1)When α›=0, the weighted least-squares regression analysis can be utilized to estimate the parameters m0 and m1, where the dependent variable is dX and the weight of independent variable is [X(1-X)]-1 Ofiteru et al. [31].
A fundamental piece of information the above listed questions
should reveal is: to what degree the neutral model could explain the dynamics of common species? For each species in the three
groups we designated previously, the purely neutral model (α’=0)
was fitted across the entire period of sampling, respectively. The
R-squared was used to measure the variability (portion of variance)
that can be explained by the neutral dynamics for each species.
With the aim to study whether or not and how the environmental
disturbances would influence the dynamics of species in micro
biota, we selected and reanalyzed a set of subsets. For individual A
(A-Gut group and A-Saliva group) who traveled abroad during the
day 71 to 122, when he moved from a major American metropolitan
area to the capital of a developing nation in Southeast Asia, where
he had rather different diet and had diarrhea during days 80 to 85
and 104 to 113. We divide the whole time period into pre-travel,
mid-travel and post-travel periods and reanalyzed A-Gut and
A-Saliva groups in 3 time periods respectively using purely neutral
model. For individual B (B-Gut group) who had an enteric infection
resulted from food poisoning induced by Salmonella sp. during the
days 151 to 159, we fitted the neutral model to the data in preinfection
and post infection respectively.
Results
Neutral dynamics of common species
After preprocessing based on the description in the Materials and Methods section, we obtained 14 species in A-Gut, 62 species in A-Saliva and 4 species in B-Gut for our analysis. To the complete time-series data of each species in 3 groups, a purely neutral model was fitted, and the results were displayed in (Tables 1, 2 & 3), respectively. The results for A-Saliva were displayed only partially in (Table 2), and the remaining was listed in the online Supplementary (Table S1). The portion of variance that can be explained by neutral model was measured by the R-squared values (R2). First, for the most common species in the two gut datasets, the portion of variance that could be explained by the neutral model is low and ranged from 0.2% to 8.1%, with the exception of Faecalibacterium prausnitzii that exhibited high neutral dynamics (R2 of model is 0.384 in A-Gut and 0.142 in B-Gut) and the most fitted models had R-squared values lower than 0.1, suggesting that neutral dynamics exerted limited effects on the gut microbial species, apart from Faecalibacterium prausnitzii. However in the saliva dataset, the R-squared values of most species were relatively high, where 25.81% (16/62) fitted models for the most common species had R-squared values higher than 0.2 and 12.9% (8/62) had R-squared values higher than 0.3, indicating that neutral dynamics may play a significant role in the saliva micro biota.
Table 1: The parameter values of the purely neutral model for the most common species in A-Gut group.
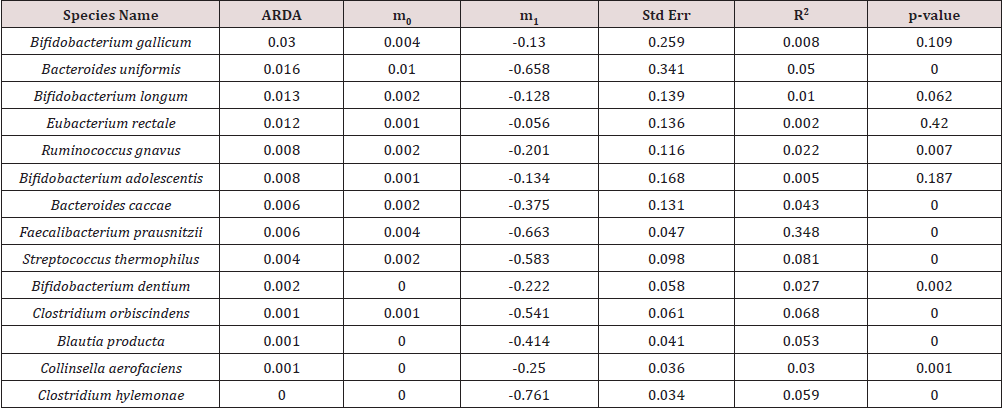
Table 2: The parameter values of the purely neutral model for the 10 most abundant most common species in A-Saliva group.
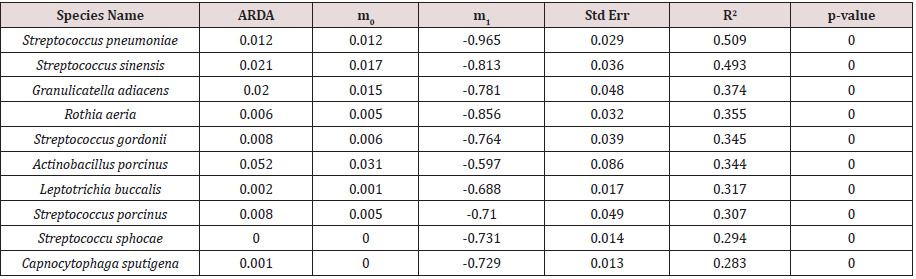
Table 3: The parameter values of the purely neutral model for the most common species in B-Gut group.
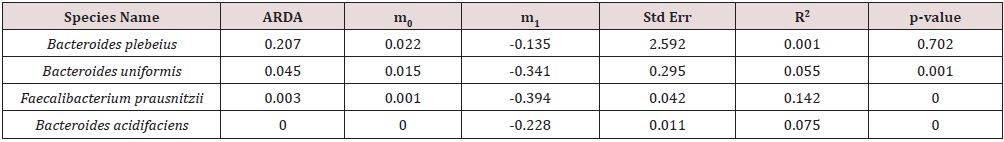
The effects of travel and infection on the neutral dynamics of common species
Individual A was exposed to new diet and had diarrhea twice
during the period of traveling, days from 71 to 122, and individual
B got infection days from 151 to 156, both of which could be
considered as dramatic environmental disturbance on internal
physiological environment. Despite the global view, our results
demonstrated that the neutral dynamics of some species were
reduced significantly by such disturbances while some others
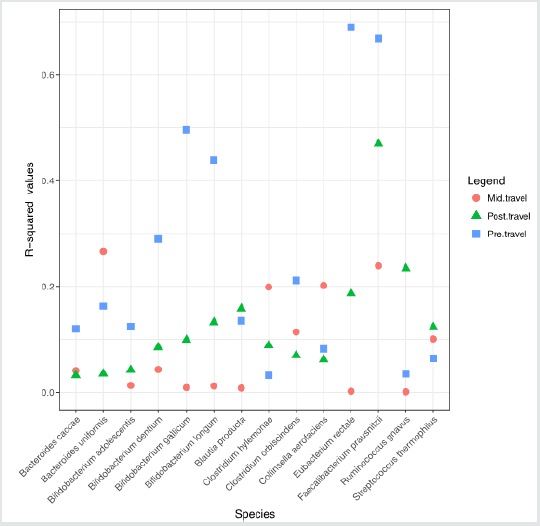
remain stable or slightly increased. In A-Gut group, for examples,
the R-squared value, which represented the effect of neutral
dynamics, for Bifid bacterium gallium dropped from 0.670 in the
per-travel to 0.005 in the mid-travel and then increased slightly to
0.100 in the post-travel, the R-squared value for Eubacterium rectal
dropped from 0.617 in the pre-travel to 0.002 in the mid-travel
and then increased slightly to 0.101 in the post-travel, and the
R-squared value for Bifid bacterium longum changed from 0.269 to
0.008 and to 0.109 finally, while the R-squared values for the other
species showed relatively very small variances.
In B-Gut group, the neutral dynamics for Bactericides plebeians
and Bactericides uniforms almost disappeared after the infection,
given that the R-squared value changed from more than 0.2 to near
0, whereas the R-squared value for Faecalibacterium prausnitzii
increased from 0.084 to 0.410, suggesting that environmental
disturbance would not always result in the loss of neutral dynamic
for some species. In the oral samples, i.e. the A-Saliva group, though
the change of variance was much more complex, the majority of
species, especially those abundant ones, stayed in the relative
high R-squared values, even after exposing to the environmental
disturbance. The curves of R-squared values for each species in
different periods in 3 groups were displayed in (Figure 1) (A-Gut),
(Figure S1) (A-Saliva) and (Figure S2) (B-Gut) and the parameters
were listed in (Table S2), (Table S3) and (Table S4) respectively.
Discussion
The relatively stable temporal dynamics of microbiota could be
a key to prevent us from the pathological disorders. However, as
mentioned in the introduction, a great deal of external and internal
factors would affect the normal microbiota and finally lead to more
susceptibility to pathogens, obesity and auto-inflammatory diseases
David et al. [35] The first step to study the long-term dynamics is
to conduct longitudinally observations on both the host and micro
biota at regular time intervals. Wet is not only the immense cost
of labor power and material resources, but also the enforcement
of individual compliance that make following the individuals for
a long time strictly a non-trivial task. Hence, the high-resolution
time-series with good quality and coupled metadata are scarce. In
David et al. [35] study, they followed two individuals daily for a year
for both micro biota and host and found micro biota can be quickly
and profoundly altered via common actions and experiences, which
made us wonder the underlying mechanism for such alteration.
From an ecological perspective, niche theory emphasizing the
deterministic factors and neutral theory underlining the stochastic
factors could be used to describe the opposite forces that shape the
micro biota. In fact, they are probably jointly responsible for the
micro biota assembly Dumbrell et al. ; Stegen et al. [32,33] From the
stationary ranked abundance distribution testing neutrality solely,
ignoring the name of taxa, the information about the dynamics of
individual taxa will be lost. However, the longitudinal dynamics of
individual taxa can be very significant in shaping the micro biota
due to specific attributions and interactions. We maintained that it
is equally important to reveal such differences.
In Ofiteru et al. [31] study, they recalibrated a neutral model to
test the neutral dynamics of individual taxa through time-series. In
Ofiteru et al. [31] study, they revealed that it is possible to explain
relative high portion of the variance in the time series of abundance
for the top two ranked taxa in two functional groups. However
they could not test all taxa due to the limitation from the T-RFLP
(Terminal Restriction Fragment Length Polymorphism) method
in failing to detect the rare taxa. We are able to test more species
using their model, thanks to the high throughput next generation
sequencing technology harnessed in David et al. study [35].
We found that relatively small portion of temporal variances of
the most common species in the gut samples could be explained
by neutral dynamics, suggesting that they have low resistances to
environmental effects. However, there are also certain species that
could exhibit high neutral dynamics in gut, such as Faecalibacterium
prausnitzii in this study. In contrast, in the saliva samples, the neutral
model could explain even higher portion of variance for amount of
common species than those taxa tested in Ofiteru et al.[31] study,
in which a well-controlled artificial environment for microbes
were sampled. One of possible reasons might be that the oral is an
open gateway linking external environment and our body, where
the environmental disturbance occurred routinely via materials exchange and foods/drinks intake, resulting in eliminations for the
species that had low resistances to disturbances and consequently
the stayed taxa could maintain high neutral dynamics.
Another merit of our study is revealing the change of
neutral dynamics for species caused by observed environmental
disturbances, which provided us a better understanding on the
underlying mechanism of the changes of individual populations
in the micro biota and a guide to better manage the important
species as well. (Table S2, S3 and S4) in the supplementary
information showed evident differences during/after the periods
of disturbances for some species. For example, in A-Gut group,
lactic acid-producing bacterium Bifid bacterium (B. gallicum and
B. longum) was found of the highest neutral dynamics, but during
the period of traveling, they lost neutral dynamics (R-squared for
neutral models dropped to near zero), which meant they would be
influenced by environmental factors more easily. Bifid bacterium is
thought to be able to inhibit the growth of pathogens and protect
human from diarrhea [36-40], and to enhance the immune function
via increase in anti-inflammatory cytokines Ouwehand et al. [41].
To some extent, for microbes, the property of neutral dynamics
may be considered as a protective factor against environmental
disturbances, hence it is possible to be associated with the dysbiosis
of certain beneficial species that the loss of neutral dynamics, such
as the example of Bifid bacterium in A-Gut, resulting from loss such
protective factor, which may be a significant reason for individual A
to get diarrhea twice during his travel period.
In the B-Gut group, two Bactericides (Bactericides plebeians
and Bactericides uniforms) were also found to loss their neutral
dynamics after the infection, though another Bactericides
(Bactericides acidifaciens) did not vary obviously in the neutral
dynamics and Faecalibacterium prausnitzii even increased
drastically. David et al. [35] found that, for individual B, more than
half of gut taxa persistently declined and did not recovery to the preinfect
state during the remaining sampling dates. Their hypothesis
that competitors replaced the lost taxa made great sense, but the
underlying ecological force was not explored. We believed that, for
some species, the loss of neutral dynamics property could increase
the possibility that they were influenced by environmental factors
and replaced by competitors, which could be one of the possible
explanations for the competitive displacement hypothesized by
David et al. [35]. In the A-Saliva samples, the condition was more
complex and most species did not experience an obvious decrease
in neutral dynamics, which is understandable given that oral has
been exposed to frequently environmental disturbances, so the left
species maintain higher resistance to environmental effects. Though
it may be too assertive to conclude that dysbiosis would occur due
to the change of neutral dynamics of some species, such influence
should not be ignored. In a big picture, such alterations probably
made the whole micro biota more fragile to environmental impacts
and pathologic dysbiosis may occur ultimately due to accumulative
effects.
Author Contributions
Y.X and Y.C conceived the study, Y.X, G.Y and Q.H analyzed the data, prepared the tables and figure and interpreter the results. Y.X wrote the paper, G.Y and Y.C reviewed the paper. All authors have read and approved the manuscript.
Competing interest
The authors declare no competing interest.
References
- Peterson J, Garges S, Giovanni M, Pamela McInnes,Lu Wang, et al. (2009)The NIH human micro biome project. GenomeResearch 19(12): 2317-2323.
- Qin J, Ruiquiang, Jeroen Raes, Jun Wang, Junming XU, et al. (2010) A human gut microbial gene catalog established by meta genomic sequencing. Nature 464(7285): 59-65.
- Licciardi PV, Zheng Quan Toh, Eileen Dunne, Sook-San Wang, Mimi Tang, et al. (2012) Protecting against Pneumococcal Disease: Critical Interactions between Probiotics and the Airway Microbiome. Plos Pathogens 8(6): e1002652.
- van Rensburg JJ,Huaiying Lin, Xiang Gao, Evelyn Toh, Qunfeng Dong, et al. (2015) The Human Skin Micro biome Associates with the Outcome of and Is Influenced by Bacterial Infection. Mbio 6(5): 01315-1500.
- Stappenbeck TS, Hooper LV, Gordon JI (2002) Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proceedings of the National Academy of Sciences USA 99(24): 15451-15455.
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JWe et al. (2005) Host-bacterial mutualism in the human intestine. Science 307(5717): 1915-1920.
- Dicksved J, Halfvarson J, Jansson JK, Rosenquist M, Tysk C, et al. (2008) Molecular analysis of the gut microbiota of identical twins with Crohn's disease. The Wesme Journal 2(7): 716-727.
- Arthur JC, Ernesto Perez- Chanona, Sarah Tomkovich, Joshua MU, Belgin Dogan, et al. (2012) Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338(6103): 120-123.
- Sun J, Kato I (2016) Gut microbiota,inflammation and colorectal cancer. Genes & Diseases 3(2):130-143.
- Le CE, Nielsen T, Qin J, Prifiti E, Falony G, et al. (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500(7464): 541-546.
- Qin J, Jung Wang, Zhiming Cai, Shenghui Li, Fan Zang, et al. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: 55-60.
- Rasiah IA, Wong L, Anderson SA, Sissons CH (2005) Variation in bacterial DGGE patterns from human saliva: Over time, between individuals and in corresponding dental plaque microcosms. Archives of Oral Biology 50(9): 779-787.
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, et al. (2013) The long-term stability of the human gut microbiota. Science 341(6141):1237439.
- Dethlefsen L, Relman DA (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences USA 108(S1): 4554-4561.
- Faa G,Garosa C, Fanni D, Nemolatto S, Fanos V,et al. (2013) Factors influencing the development of a personal tailored microbiota in the neonate, with particular emphasis on antibiotic therapy. The journal of maternal-fetal & neonatal medicine: The official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstetricians 26(S2): 35-43.
- Korpela K, Salonen A, Virta LJ, Bork P, Kekkonen RA, et al. (2016) Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nature Communications 7: 10410.
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, et al. (2009) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine 1(6): 6ra14.
- Dash S, Clarke G, Berk M, Jacka FN (2015) The gut microbiome and diet in psychiatry: Focus on depression. Current Opinion in Psychiatry28(1): 1-6.
- Sekirov I, Russell SL, Antunes LC, Finlay BB (2010) Gut microbiota in health and disease. Physiological Reviews 90(3): 859-904.
- Ventura Met al. (2014) The gut microbiota in health and disease. Human Microbiota & Microbiome
- VandermeerJH(1972) Niche Theory. Annual Review of Ecology and Systematics 3: 107-132.
- Hirzel AH, Lay GL (2008) Habitat suitability modelling and niche theory. Journal of Applied Ecology 45: 1372-1381.
- Hubbell SP (2001) The Unified Neutral Theory of Biodiversity and Biogeography, Princeton University Press pp. 392.
- Alonso D, Etienne RS, Mckane AJ (2006) The merits of neutral theory. Trends in Ecology & Evolution 21(8): 451-457.
- Mcgill BJ, Maurer BA, Weiser MD (2006) Empirical evaluation of neutral theory.Ecology 87(6): 1411-1423.
- Rosindell J, Hubbell SP, He F, Harmon LJ, Etienne RS, et al.(2012) The case for ecological neutral theory. Trends in Ecology & Evolution27(4): 203-208.
- Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, et al. (2016) Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. The WeSME Journal 10(3): 655-664.
- Leibold MA, McPeek MA (2006) Coexistence of the niche andneutralperspectivesincommunityecology. Ecology 87(6): 1399-1410.
- Chave J (2004) Neutral theory and community ecology. Ecology Letter 7(3): 241-253.
- Ofiteru ID, Mary Lunn, Thomas P Curtis, Craig S Criddle, William T. Sloan, et al. (2010) Combined niche and neutral effects in a microbial wastewater treatment community. Proceedings of the National Academy of Sciences of the United States of America 107(35): 15345-15350.
- Dumbrell AJ,Nelson M, Helgason T, Dytham C,Fitter AH, et al. (2010) Relative roles of niche and neutral processes in structuring a soil microbial community. The ISME Journal 4(3): 337-345.
- Stegen JC, Xueju Lin, Konopka AE, Fredrickson JK (2012) Stochastic and deterministic assembly processes in subsurface microbial communities. The ISME Journal 6: 1653-1664.
- Wells GF, Park HD, Yeung CH, Eggleston B, Francis CA,et al.(2009) Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: Betaproteobacterial dynamics and low relative abundance of crenarchaea. Environmental Microbiology 11(9): 2310-2328.
- David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, et al. (2014) Host lifestyle affects human microbiota on daily timescales. Genome Biology 15(7): R 89.
- Saavedra JM, Bauman NA, Oung I, Perman JA, Yolken RH, et al.(1994) Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhea and shedding of rotavirus. Lancet 344(8929): 1046-1049.
- Shu Q, Qu F, Gill HS (2001) Probiotic treatment using Bifidobacterium lactis HN019 reduces weanling diarrhea associated with rotavirus and Escherichia coli infection in a piglet model. Journal of Pediatric Gastroenterology & Nutrition 33(2): 171-177.
- Yuan J, Zhu L, Zhang Y, Ying T, Wang B, et al. (2006) A proteome reference map and proteomic analysis of Bifidobacteriumlongum NCC2705. Molecular & Cellular Proteomics 5(6): 1105-1118.
- İşlek A, Sayar E,Yılmaz A,Baysan BÖ, Mutlu D, et al. (2014) The role of Bifidobacterium lactis B94 plus inulin in the treatment of acute infectious diarrhea in children. Turkish Journal of Gastroenterology the Official Journal of Turkish Society of Gastroenterology 25(6): 628-633.
- Mercenier A, Blum-Sperisen S, Rochat F (2014) Use of Bifidobacterium longum for the prevention and treatment of inflammation: US, US8916145.
- Ouwehand AC, Bergsma N, Parhiala R, Lahtinen S, Gueimonde M, et al.(2008) Bifidobacterium, microbiota and parameters of immune function in elderly Subjects. FemsWemmunology& Medical Microbiology 53(1): 18-25.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...