
Lupine Publishers Group
Lupine Publishers
Menu
Research Article(ISSN: 2641-6875) 
Formation of Cell “Nest” in Cultured Synthetic DNA (Streptomyces) Crown Cells-Microscopic Appearance of Cells Within “Nests Volume 2 - Issue 2
Shoshi Inooka*
- Japan Association of Science Specialists, Japan
Received: March 27, 2023; Published: April 04, 2023
*Corresponding author: Shoshi Inooka, Japan Association of Science Specialists, Japan
DOI: 10.32474/CTBM.2023.03.000166
Abstract
aminonitriles is a biosynthetic that allows researchers to expand their horizons and identify promising therapeutic potential. This study aims to evaluate the antibacterial, antifungal and antiparasitic potential of aminonitrils. The methodology adopted was a bibliographic review. The guiding question was elaborated according to the PICo strategy. The databases consulted were PubMed, BVS and Elsevier, using the descriptors: “aminonitriles”, “amino nitriles”, “antifungals”, “antibacterial”, “antimicrobial activity”, “antiprotozoals” and “antiparasitic agents”. The titles and abstracts were read, the selected ones were approached in full. Descriptive studies of the role of aminonitriles in antimicrobial agents were elected. The work with drugs demonstrates broad bioactive activity, covering bacteria, fungi and protozoa, of these, some species have values found close to the standard of combat for the strains in question. It was observed, in the literature, relevant activity against the bacteria Staphylococcus aureus, Escherichia coli, Salmonella Typhi, Mycobacterium tuberculosis; strains of the fungi Candida neoformans, Candida albicans and against the protozoan Trypanosoma cruzi. Furthermore, after a bibliographical analysis, the antitumor potential of aminonitrils was noted. The data obtained reveals an alternative perspective from the point of view of the scenario of infection by microorganisms, which can support future research on the pharmacological effect of aminonitriles.
Keywords: Aminonitriles; antibiotics; biosynthetic
Introduction
DNA crown cells are artificial cells that are covered with DNA [1-3]. They are generated by incubating synthetic DNA crown cells in egg white and can be prepared using a mixture of sphingosine (Sph)-DNA and adenosine-monolaurin (A-M) compounds [4,5]. Synthetic DNA (Escherichia coli) crown cells have been shown to form assemblies spontaneously in the presence of commercial salts [6], inorganic solutions, [7], Bacillus subtilis [8,9], and yeast [10]. Most such assemblies changed into crystal-like substances after treatment with monolaurin. In spontaneously formed assemblies, cells proliferated after two treatments with monolaurin [11]. When MgCl2 was used, assemblies were formed without two monolaurin treatments [12]. The previous experiments were conducted using synthetic DNA (E. coli) crown cells. However, it is unclear whether cell proliferation can occur in other synthetic DNA crown cells. The present study therefore examined whether cell proliferation occurred in cultured synthetic DNA (Streptomyces) crown cells.
Materials and Methods
Materials
The following materials were used: Sph (Sigma, USA and Tokyo Kasei, Japan), DNA (from the mixtures of Streptomyces griseus and Streptomyces kanamyceticus), adenosine (Sigma and Wako, Japan), monolaurin (Tokyo Kasei), and MgCl2 (Hayashi Junyaku Co. Ltd., Japan). In addition, a compound synthesized from A-M was used. Monolaurin solutions were prepared to a concentration of 0.1 M in distilled water. MgCl2 solutions were prepared to a concentration of 20% in distilled water.
Methods
Preparation of Synthetic DNA (Streptomyces) Crown Cells
The generation of synthetic DNA (Streptomyces) crown cells using Sph-DNA-A-M was performed as described previously. Briefly, 180 μL of Sph (10 mM) and 90 μL of DNA (1.7 μg/μL) were combined, and the mixture was heated and cooled, twice. A-M (100 μL) was added, and the mixture then was incubated at 37°C for 15 min. Next, 30 μL of monolaurin was added, and the mixture was incubated at 37°C for another 5 min. The resulting suspension was used as synthetic DNA (Streptomyces) crown cells.
Cell Proliferation Tests
a) Twenty-five microliters of synthetic DNA (Streptomyces) crown cells was incubated with and without 25 μL of MgCl2 solution for 18 h at 37°C.
b) Twenty-five microliters of monolaurin were added to the synthetic DNA (Streptomyces) crown cells.
c) The mixture was incubated for approximately 1 and 2 days at 37°C.
d) The samples prepared in step 3 were observed under a light microscope.
Microscopic Observations
A drop of each sample was placed on a slide glass and covered with a cover glass. The resulting slide was then observed under a light microscope.
Results and Discussion
a) Microscopic appearance of synthetic DNA (Streptomyces) crown cells cultured for 18 h without MgCl2 and incubated with monolaurin for 1 day. Figure 1 shows the microscopic appearance of cultured DNA (Streptomyces) incubated in monolaurin for 1 day. Various floating objects of different sizes and shapes were observed (Figure1 arrows a-d). Such objects may be large cells, but their size could not be determined. On the other hand, it is well known that “nests” are places where animals such as mice are born and they make nest within which to bear their young. Since the structures described here might be the site of cell generation and cell mature, these objects are referred to here as “cell nests” or simply “nests”. A round cell (Figure 1 arrow e) measuring approximately 3-4 μm was observed within a nest (Figure1 arrow a). Figure 2 (arrow a) shows the magnification of the micrograph in Figure 1 (arrow a). The micrograph shows a nest that was constricted in the middle (arrow b) and that contained two clear cells on one side (arrow c). As described previously [13], because color was easy to observe in the micrographs similar substances could not be discerned, only different substances The net (equivalent to outer membrane) of nest in Figure2 (arrow a) consists of at least two layers, a yellowgreen color layer and an orange layer. Figure 3 shows other cultured synthetic DNA (Streptomyces) crown cells that were incubated with monolaurin for 1 day. Two nests which were constricted (arrow c) and contained cells were observed (arrows a and b). A round nest containing cells was observed (arrow d). Also, a nest without cells was observed (arrow e). The approximate cell size was 2–3 μm (arrow f). Figure 4 shows the magnification of the micrograph shown in Figure 3 (arrows a, d and e). A nest (arrow a) was constricted in one side (arrow d) and cells were contained in that side of the nest (arrow e). One nest (arrow b) contained cells, but another nest (arrow c) did not contain cells. A nest in Figure 4 (arrow a) is divided into two nests at the point of constriction, forming two nests (arrows b and c).
b) Microscopic appearance of synthetic DNA (Streptomyces) crown cells incubated for 18 h without MgCl2 and followed incubation with monolaurin for approximately 2 days. Numerous nests were observed (Figure 5), some of which contained cell-like structures (arrows a, b and c) that measured approximately 3-4 μm (arrow d). Figure 6 shows the magnification of the micrograph shown in Figure 5 (arrow c). A nest (arrow a) contained about 10 cells of various sizes (arrows b and c). Figure 7 (arrow a) shows the magnification of the micrograph shown in Figure 5 (arrow d). Many cells of various sizes and shapes were observed within the nest (arrow b). The nets of nests were degraded (arrows c and d) and cells within the nest protruded from the nest (arrow e). Figure 8 shows another picture of a synthetic DNA (Streptomyces) crown cell incubated without MgCl2 and in monolaurin for 2 days. A total of four nests were observed (arrows a, b, c and d), with three nests containing many cells (arrows a, b and c); one nest (arrow d) containing several cells (arrow d). The size of cells within nests was approximately 2–3 μm (arrow e). Figure 9 (arrow a) shows the magnification of the micrograph shown in Figure 8 (arrow b). Many cells of various sizes and shapes were observed (arrow c). Degradation of the net of the nest (arrow b) resulted in cells escaping from the nest (arrow d).
c) Microscopic appearance of synthetic DNA (Streptomyces) crown cells incubated with MgCl2 for 18 h and then with monolaurin for 2 days. Many nests of various sizes and shapes were observed; these are shown in Figure 10 (arrows a-f, m&h). Every nest contained cell-like structures. Constrictions in nests (arrows a, b and c) were observed and cells were seen outside the nests (arrow j). The approximate cells size was 2-3 μm (arrow g). Figure 11 shows a magnification of the micrograph shown in Figure 10 (arrows a, i and j) Cells of various sizes were observed (Figure 11, arrows d, e f and k). Among these cells, some had the appearance of very small dots (arrows d and e), whereas others (arrows f and k) were clear and round. The findings suggest that the cells shown in Figure 11 (arrows d and c) had just been generated, whereas the cells shown by arrows f and k had the appearance of mature cells. A narrow nest was observed, and a net of a nest appeared degraded (Figure 11 arrow b). Many cells were observed outside of nest (Figure 11 arrow a) and most of these cells were protruding from the nest (Figure 11 arrow c).
Figure 1: Microscopic appearance of cultured synthetic DNA (Streptomyces) crown cells after 18 h without MgCl2 followed by incubation in monolaurin for 1 day. Free-floating objects of various sizes and shapes (a, b, c and d). Round cells (e) measuring approximately 3–4 m within a nest in (a).
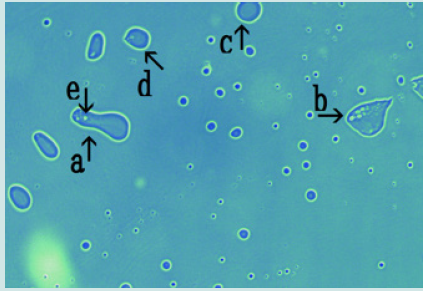
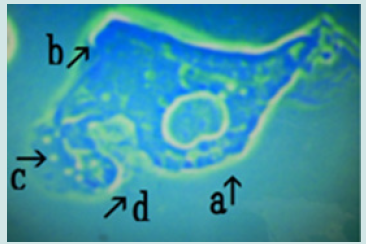
Figure 2: Magnified appearance of features shown in the micrograph of Figure 1 (arrow a). A nest (arrow a) constricted in the middle (arrow b) containing two cells at one side was observed (arrow c).
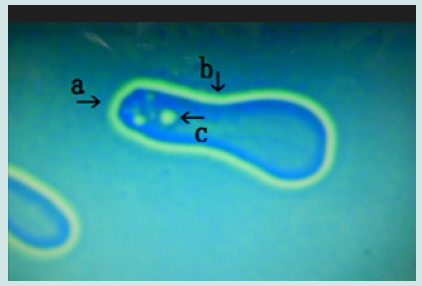
Figure 3: Microscopic appearance of another cultured synthetic DNA (Streptomyces) crown cell incubated with monolaurin for 1 day. Two nests were constricted (arrow c) and contained cells (arrows a and b). A round nest containing cells (arrows d) and a nest without cells (arrow e). Approximate cell size is 2–3 m (arrow f).
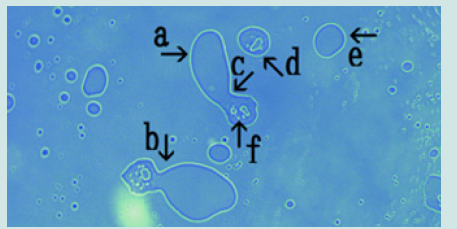
Figure 4: Microscopic appearance of the micrograph in Figure 3 (arrows a, d and e). A nest (arrow a) is constricted at one side (arrow d) and contains cells (arrow e). A nest containing cells (arrow b). A nest with no cells (arrow c).
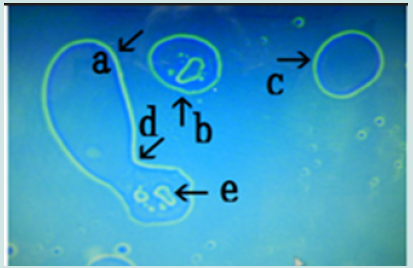
Figure 5: Microscopic appearance of synthetic DNA (Streptomyces) crown cells incubated for 18 h without MgCl2 followed by incubation with monolaurin for approximately 2 days. Nests containing cell-like structures (arrows a, b and c). Approximate cell size was 3–4 m (arrow d).
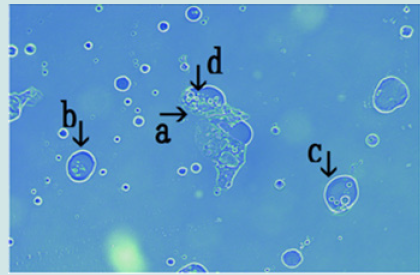
Figure 6: Microscopic appearance of the micrograph shown in Figure 5 (arrow c). Nest (arrow a) contains about 10 cells of various sizes (arrows b and c).
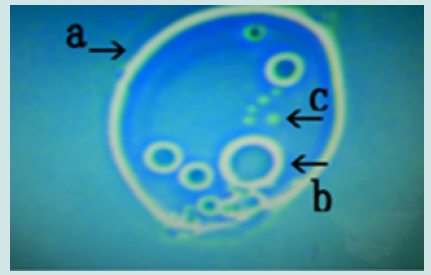
Figure 7: Microscopic appearance of the micrograph shown in Figure 5 (arrow d). Many cells of various sizes and shapes are shown within nest (arrow b). The nets (equivalent to outer membrane) in the nest were destroyed (arrows c and d) and cells within the nest protruded from the net (arrow e).
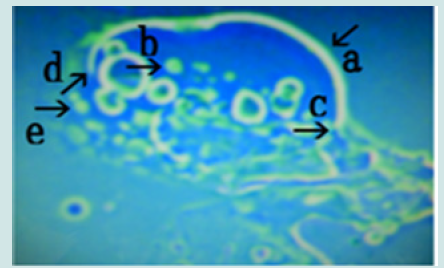
Figure 8: Microscopic appearance of another synthetic DNA (Streptomyces) crown cell incubated without MgCl2 and with monolaurin for 2 days. Four nests are shown (arrows a, b, c and d). Three nests contain many cells (arrows a, b and c) and one nest contains several cells (arrow d). Each cell within a nest (arrow e) measure approximately 2–3 m.
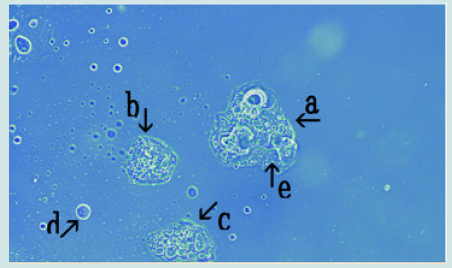
Figure 9: Microscopic appearance of the micrograph shown in Figure 8 (arrow b). Cells of various sizes and shapes (arrow c). Rupture of the net of the nest (arrows a and b) resulting in the escape of cells from the nest (arrow d).
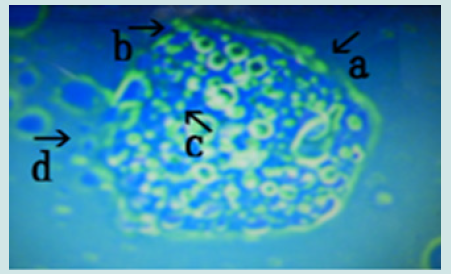
Figure 10: Microscopic appearance of synthetic DNA (Streptomyces) crown cells incubated in MgCl2 for 18 h and then with monolaurin for 2 days. Nests of various sizes and shapes (arrows a, b, c, d, e, f and h). All nests contain cell-like structures. Narrowing of nests (arrows a, b and c) and cells outside nests (arrow j). Approximate cell size is 2–3 m (arrow g).
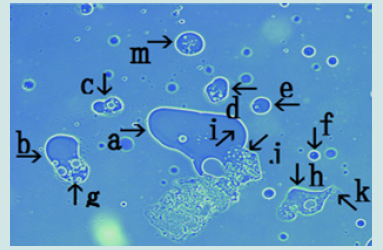
Figure 11: Microscopic appearance of the micrograph shown in Figure 10 (arrows a, i and j) of cells of varying sizes (arrows d, e, f and k) within a nest (arrow a). Narrowing of a nest and the net of a nest (arrow b). Many cells are outside the nest, and many cells protrude from the nest (arrow c).
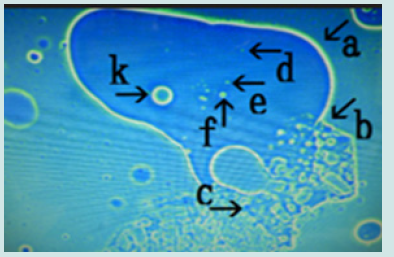
The findings showed that a large number of cells were formed in a nest within 2 days of culture with monolaurin and that the cells were released from the nest. Figure 12 shows the magnification of the micrograph in Figure 10 (arrows b and g). Narrowing of the nest (arrow a) was observed (arrow b) and many cells in various sizes were observed outside of the nest (arrow c). Figure 13 (arrow a) shows magnification of the micrograph shown in Figure 10 (arrow c). Cells of various sizes (arrow b) were observed within nest (arrow a). The net of the nest was degraded (arrow c). Figure 14 shows the magnification of the micrograph in Figure 10 (arrow d). Numerous cells (arrow b) were observed within the nest (arrow a). Figure 15 shows the magnification of the micrograph in Figure 10 (arrow e). Four cells (arrows b, c, d and e) were observed within the nest (arrow a). Figure 16 shows the magnification of the micrograph in Figure 10 (arrow f). The nest (arrow a) contained a cell (arrow b). Figure 17 shows the magnification of the micrograph in Figure 10 (arrow m).
More than 15 cells (arrow b) were observed within the nest (arrow a). Figure 18 shows a magnification of the micrograph in Figure 10 (arrows h and k). The net of the nest (Figure 18 arrow a) was degraded (arrows b and d) and cells within nest were escaped from the nest (arrow c). The present study examined whether cell proliferation, previously observed using synthetic DNA (E. coli) crown cells, was also observed using synthetic DNA (Streptomyces) crown cells. In synthetic DNA (E. coli) crown cells, cell proliferation occurred from spontaneously formed assemblies in samples that were treated twice with monolaurin after incubation in monolaurin for approximately 2 days. Also, in an experiment in which MgCl2 was added to the culture medium, cell proliferation occurred after incubation with monolaurin for approximately 2 days, without being stimulated twice. These examples were cell proliferation from the assemblies that were formed with synthetic DNA (E. coli) crown cells, and which proliferated spontaneously or after the addition of MgCl2. On the other hand, cell occurrence was spontaneously observed in nests that formed during the cultivation of synthetic DNA (Streptomyces) crown cells, in the present study. In the cultures using MgCl2, several kinds of cell populations were observed, including cell populations previously reported using synthetic DNA (E. coli) crown cells.
Figure 12: Microscopic appearance of synthetic DNA (Streptomyces) crown cells incubated in MgCl2 for 18 h and then with monolaurin for 2 days. Nests of various sizes and shapes (arrows a, b, c, d, e, f and h). All nests contain cell-like structures. Narrowing of nests (arrows a, b and c) and cells outside nests (arrow j). Approximate cell size is 2–3 m (arrow g).
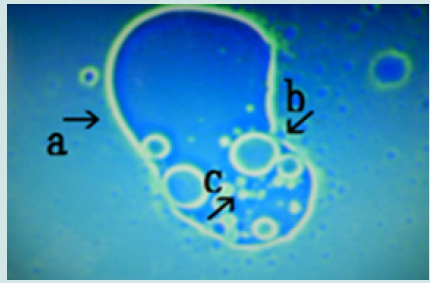
Figure 13: Microscopic appearance of the micrograph shown in Figure 10 (arrows a, i and j) of cells of varying sizes (arrows d, e, f and k) within a nest (arrow a). Narrowing of a nest and the net of a nest (arrow b). Many cells are outside the nest, and many cells protrude from the nest (arrow c).
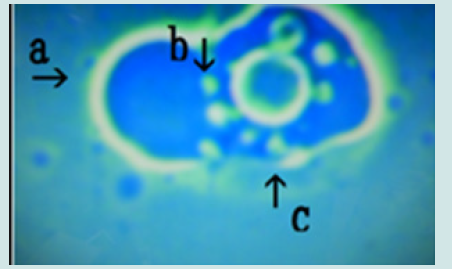
Figure 14: Microscopic appearance of synthetic DNA (Streptomyces) crown cells incubated in MgCl2 for 18 h and then with monolaurin for 2 days. Nests of various sizes and shapes (arrows a, b, c, d, e, f and h). All nests contain cell-like structures. Narrowing of nests (arrows a, b and c) and cells outside nests (arrow j). Approximate cell size is 2–3 m (arrow g).
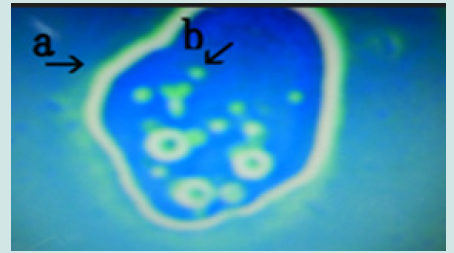
Figure 15: Microscopic appearance of the micrograph shown in Figure 10 (arrows a, i and j) of cells of varying sizes (arrows d, e, f and k) within a nest (arrow a). Narrowing of a nest and the net of a nest (arrow b). Many cells are outside the nest, and many cells protrude from the nest (arrow c).
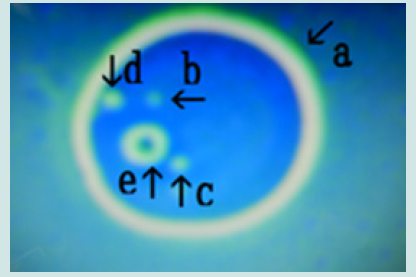
Figure 16: Microscopic appearance of synthetic DNA (Streptomyces) crown cells incubated in MgCl2 for 18 h and then with monolaurin for 2 days. Nests of various sizes and shapes (arrows a, b, c, d, e, f and h). All nests contain cell-like structures. Narrowing of nests (arrows a, b and c) and cells outside nests (arrow j). Approximate cell size is 2–3 m (arrow g).
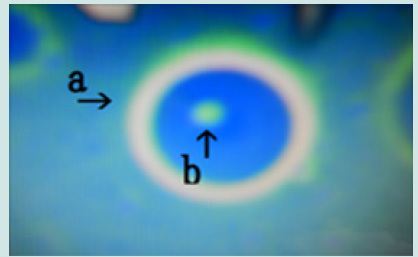
Figure 17: Microscopic appearance of the micrograph shown in Figure 10 (arrows a, i and j) of cells of varying sizes (arrows d, e, f and k) within a nest (arrow a). Narrowing of a nest and the net of a nest (arrow b). Many cells are outside the nest, and many cells protrude from the nest (arrow c).
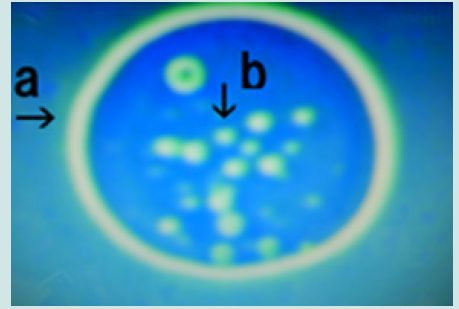
Figure 18: Microscopic appearance of the micrograph shown in Figure 10 (arrows a, i and j) of cells of varying sizes (arrows d, e, f and k) within a nest (arrow a). Narrowing of a nest and the net of a nest (arrow b). Many cells are outside the nest, and many cells protrude from the nest (arrow c).
Cell populations within nests are reported here for the first time. Therefore, the present findings showed that there were at least two ways in which cells can derive: cell proliferation from assemblies and cell formation within nests. The most interesting question regarding the present study is how the cells were born within nests. In cells that arise in nests, two hypotheses can be proposed. One is that nests enclose ready-made synthetic DNA (Streptomyces) crown cells or related particles that matured within the nest; indeed, this development appears to have been demonstrated by microscopic findings. It has already been demonstrated that particles (Sph- DNA-adenosine) were formed and that cells were formed from these particles within egg white. So, such particles may be enclosed within nests and cells may be synthetically born from these particles. Thus, it appears that cells or particles already exist within nests and proceed to multiply within nests. Another hypothesis is that the materials that formed cells exist within nests and cells are then synthesized within nests. It has already been demonstrated that synthetic DNA (E. coli) crown cells formed crystal-like substances after treatment with monolaurin and that most cells disappeared. Therefore, it is considered that all of the materials, including cells, that were prepared in the start of experiments were reconstructed within nests. New cells may be generated from these materials within nests.
In summary, cells, including their component materials (sphingosine, DNA, adenosine and monolaurin) which comprise synthetic DNA crown cells were used to reconstruct cells within a nest and that new cells form from these materials. On the other hand, it was clearly shown that cell occurrence was observed within nests and many micrographs of nests showed that cell occurrence occurred. However, at present, details of nest formation, scale, function, the components and so on, are not clear. Indeed, there is very little evidence related to the occurrence of cells and their formation within the nest. In terms of cell occurrence, as shown in Figure 11, cells the size of small dots (arrows e and d) and rings (arrows f and k) were observed. Cells like dots may be generated, but this may occur only after they appear as rings. However, it was unclear whether dot-like cells (arrows e and d) transform into ringlike cells (arrows f and k). In particular, the ring-like cells in Figure 6 (arrow b) and Figure 11 (arrow k) were observed in many nests, as shown in other (Figures 7, 12-15). Therefore, it was unclear whether such ring-like cells grew from dot-like cells; it is possible that could be formed in other ways. After many cells occur within a nest, the nest appears to degrade and all of the cells within the nest are released, as shown in Figure 7.
The findings showed that a large number of cells were produced in this way. Although there is no evidence, it may be possible that the ring-like cells shown in Figure 6 (arrow b) and Figure 11 (arrow k) developed into nests which then formed new cells. On the other hand, although cells are formed and multiply in nests, at present, no definitions for cell nests exist. Nests of various sizes are shown in Figure 10. The nest in Figure 10 (arrow a) was the largest, whereas that shown by arrow f was the smallest; nests shown by arrows b, c, d, and e were intermediate in size. It is proposed that in nest formation, first, larger nests (e.g., those shown by arrows a or b) may be formed. Then, these nests divide into two nets to form nests like those shown by arrows d and e. However, it is unclear whether such nests formed based on established biological principles or whether their formation was random. As shown in Figures 15- 17, the nets of nests are yellow-green and orange in color. When culturing synthetic DNA (E. coli) crown cells (13), yellow-green objects are called outside structures (o-structures) and the orange spaces are called middle structures (m-structures). Following this nomenclature, the nets of nests are considered to consist of o-structures and m-structures. Such substances may consist of Sph, DNA, adenosine and monolaurin, which were used to construct the synthetic DNA crown cells. However, it is unclear whether these nets are composed of materials such as Sph or DNA.
Conclusion
In the present experiments, cultured synthetic DNA (Streptomyces) crown cells were used and nest formation was observed within 2 days of cultures. On the other hand, no nests were observed, or they may have been overlooked in the experiments of the synthetic DNA (E. coli) crown cells which proliferated from assemblies. Thus, these observations may be a characteristic of cell occurrence in synthetic DNA crown cells. Thus, the present experiments on the use of synthetic DNA (Streptomyces) crown cells demonstrated that nests were formed within 1 day after treatment with monolaurin and that a large number of cells were formed within nests and then released from the degraded nest. Further experiments will be required to clarify whether cells that developed in nests and which were then released can grow at another site (nest), as in metastasis. Also, these cells may behave like a virus that multiplies within cells and is then released into the environment from where they live in other hosts (nests) and are biologically active [14,15].
References
- Inooka S (2012) Preparation and cultivation of artificial cells. App Cell Biol 25: 13-18.
- Inooka S (2016) Preparation of Artificial Cells Using Eggs with Sphingosine-DNA. J Chem Eng Process Technol 7(1): 277.
- Inooka S (2016) Aggregation of sphingosine-DNA and cell construction using components from egg white. Integr Mol Med 3(6): 1-5.
- Inooka S (2017) Biotechnical and Systematic Preparation of Artificial Cells. Glob J Res Eng 17(C1): 1-10.
- Inooka S (2017) Systematic Preparation of Artificial Cells (DNA Crown Cells). J Chem Eng Process Technol 8: 327.
- Inooka S (2022) Assemblies Formation of Synthetic DNA (E.coli) Crown Cells with Salt. “Reconstruction and Regeneration of Synthetic DNA Crown Cells”. Am J Biomed Sci Res 16(1): 1-6.
- Inooka S (2022) The assembly in synthetic DNA crown cells with inorganic salts and the transformation of DNA crown cells to crystal like substances in the presence of monolaurin. Nov Res Sci 11(2): 000759.
- Inooka S (2021) Assembly and Bacterial Growth Suppression in the Co-cultures of Synthetic DNA (Akoya Pearl Oyster) Crown Cells with Bacillus subtilis. Applied Cell Biology Japan 34: 67-84.
- Inooka S (2022) Assemblies in Synthetic DNA (Nannochloropsis Species) Crown Cells with Bacillus subt. Acta Sci Microbiol 5(1): 3-8.
- Inooka S (2021) Synthetic DNA Crown Cells Kill Yeast: Relationship with Assembly Formation of Yeast. Civ Eng Res J 12(2): 555831.
- Inooka S (2022) Cell Proliferation from the Assembly of Synthetic DNA ( Coli) Crown Cells with Stimulation by Monolaurin Twice. Curr Trends Biotechnol Microbiol 2(5): 493-498.
- Inooka S (2023) Cell Proliferation in Assemblies of Synthetic DNA ( coli) Crown Cells in the Presence of MgCl2]. Curr Trends Biotechnol Microbiol 3(3): 577-581.
- Inooka S (2022) Microscopic appearance of synthetic DNA ( coli) crown cells in primary culture. App Cell Biol Japan 35: 71-98.
- Inooka S (2022) Microscopic Appearance of Synthetic DNA ( coli) Crown Cells in Secondary Cultures. Nov Res Sci 12(4): 1-6.
- Inooka S (2022) Preparation of a DNA ( coli) Crown Cell line in Vitro-Microscopic Appearance of Cells. Ann Rev Res 8(1): 1-6.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...





