
Lupine Publishers Group
Lupine Publishers
Menu
Research Article(ISSN: 2641-6875) 
Bacteriocins Production Potentials of some Gram Positive Cocci Bacteria Isolated from Milk and other Food Sources Volume 2 - Issue 2
Tyokusa AG1* and Owuama CI2
- 1Microbiology Department, Joseph Sarwuan Tarka University, Makurdi, Nigeria
- 2Microbiology Department, Modibbo Adama University, Yola, Nigeria
Received: June 19, 2023; Published: June 30, 2023
*Corresponding author: Tyokusa AG, Microbiology Department, Joseph Sarwuan Tarka University, Makurdi, Nigeria
DOI: 10.32474/CTBM.2023.03.000172
Abstract
Bacteria isolated from six different sources viz; cow milk, goat milk, sheep milk, burukutu, dawadawa and kunu- zaki were identified using Vitek 2 compact system. The identified isolates are Staphylococcus lentus, Staphylococcus pseudintermedius, Staphylococcus sciuri, Kocuria kristinae, Kocuria rosea, Kocuria rhizophila, Enterococcus faecium, Enterococcus gallinarum, Enterococcus casseliflavus, Granulicatella elegans and Pediococcus pentosaceus. Out of these isolates, Enterococcus faecium, Enterococcus gallinarum, Enterococcus casseliflavus, Granulicatella elegans and Pediococcus pentosaceus are lactic acid bacteria. The agar well diffusion method was used in testing the bacteriocins production potentials of all the isolates using Staphylococcus aureus, Listeria monocytogenes, Salmonella typhi and Escherichia coli as test organisms. Results showed that 20 out of the 23 isolates had antimicrobial activities. Isolate G8 (from goat milk) produced the highest zones of inhibition of 23mm against Listeria monocytogenes and 22mm against Staphylococcus aureus. B2 (isolate from burukutu) was the only isolate that inhibited all the test organisms. All the isolates produced acid though at varying degrees.
Keywords: Bacteriocin; vitek 2; gram positive cocci; lactic acid bacteria; kocuria
Introduction
Bacteriocins are ribosomally synthesized antimicrobial peptides produced by bacteria that are capable of killing or inhibiting bacteria strains closely related or non-related to producing bacteria but will not harm the bacteria themselves due to their possession of specific immunity proteins [1]. Bacteriocins are capable of controlling clinically relevant susceptible and drug-resistance bacteria [2]. The vast majority of all bacteria species appear to be able to produce at least one bacteriocin, most of which are unknown. Bacteriocins producing bacteria could be extracted from various sources such as water, soil, animal intestines and food products. Bacteriocins may be secreted by both Gram positive and Gram negative bacteria. In particular, among Gram positive bacteria, bacteriocins derived from lactic acid bacteria are of great interest [3]. Reports on the first bacteriocin production were from Escherichia coli in 1925, and this peptide was named “colicins” to reflect the microbial source [4]. Bacteriocins are named according to the group of the producing bacteria, thus pediocins are produced by Pediococci sp [5], staphylococcins produced by Staphylococci sp [6], enterocins produced by Enterococci [7,8].
In recent years, a group of antibacterial proteins produced by Gram positive bacteria have attracted great interest in their potential use as food preservatives and as antibacterial agents to combat certain infections due to Gram positive pathogenic bacteria [9]. Bacteriocins of Gram positive bacteria are as abundant and even more diverse as those found in Gram negative bacteria. Firstly, bacteriocin production is not necessarily lethal to the producing cell due to the transport mechanisms Gram positive bacteria encode to release bacteriocin toxin. Typically, their biosynthesis is self-regulated with specifically dedicated transport mechanisms facilitating release, although some employ the sec-dependent export pathway. Secondary, the Gram positive bacteria have evolved bacteriocin-specific regulation, whereas bacteriocins of Gram negative bacteria rely solely on host regulatory networks [10]. Given that bacteriocins are produced by both Gram positive and Gram negative bacteria from diverse sources, this work therefore, examines Gram positive cocci isolated from different milk and food sources for their ability to produce bacteriocins against Staphylococcus aureus, Listeria monocytogenes, Salmonella typhi and Escherichia coli.
Methodology
Isolation and Identification
Fresh cow milk, goat milk, sheep milk, dawadawa (fermented cooking condiment made from locust beans seeds), burukutu (a locally prepared alcoholic beverage made from guinea corn grains) and kunu- zaki (a locally prepared non-alcoholic beverage made from guinea corn grains) were collected in sterile containers and taken to the laboratory and allowed to ferment for 24 hours. Serial dilutions were made for all the samples and appropriate dilutions (10-5 to 10-9) inoculated on MRS agar (Dextrose-20, Agar 12., Proteose peptone-10, Beef extract-10, Yeast extract-5, Sodium acetate-5, Ammonium citrate-2, Dipotassium phosphate-2, Tween 80-1, Magnesium sulphate-0.10 and Manganese sulphate-0.05 all g/L/, pH- 6.5 at 250C) TITAN BIOTECH LTD India for 24 to 48 hours at room temperature (30-370C). Discrete colonies observed were sub-cultured on MRS agar to obtain pure isolates. The pure isolates were Gram stained and the Gram-positive cocci bacteria isolates were examined further. The Gram positive cocci bacteria isolates were further identified using Vitek 2 compact system (BIOMERIEUX , France).
Four test organisms comprising two Gram positive bacteria (Staphylococcus aureus and Listeria monocytogenes) and two Gram negative bacteria (E. coli and Salmonella typhylum) were used to test for the bacteriocins production abilities of the isolates. For the preparation of crude bacteriocins, 10ml of MRS broth was inoculated with 0.1ml (McFarland standard of 0.5) of freshly prepared cultures of each isolates and then incubated at 370C for 24 hr. The 24 hr cultures were centrifuged at 6000rpm for 20min. The supernatants were filtered using cellulose acetate paper of pore size of 0.02μm to obtain cell free extracts [11,12]. The pH of the extract was adjusted to 5.5 with 1% sodium hydroxide solution. To determine the bacteriocin activity, agar well diffusion method was used. The four test organisms were then inoculated on Muller Hilton agar (Merck, Germany) and a sterile cork borer (6mm diameter) was used to bore holes on the agar surfaces. Consequently, 50μl of the extract was pipetted into the appropriate holes and allowed to absorb, then incubated for 24 hours at 370C [13,14]. After 24 hr, zones of inhibition where present were measured and recorded in millimeter (mm) [15]. To test for the acid producing abilities, the isolates were grown in MRS broth with an initial pH of 6.2 and incubated at room temperature for 5 days. The pH of each isolate in the broth was measured with a pH metre (pHep HANNA instruments) at 24 hourly intervals, i.e. 24, 48, 72, 96 and 120 hr.
Results
Results of Gram-positive cocci isolated from cow milk, goat milk, sheep milk, dawadawa, burukutu and kunu-zaki is shown in the table below. KEY: B2 and B3 (Burukutu isolates); C2, C4, C9 and C10 (Cow milk isolates); D3, D5 and D8 (Dawadawa isolates); G1, G2, G4, G6, G7 and G8 (Goat milk isolates); K1, K3, K4, K5 and K6 (Kunu- zaki isolates and S1, S2 and S5 (Sheep milk isolates) Table 1 above contained 23 Gram positive cocci consisting of Staphylococcus pseudintermedius (4), Staphylococcus lentus (3), Staphylococcus sciuri (1), Kocuria kristinae (3), Kocuria rhizophila (1), Kocuria rosea (1), Enterococcus faecium (2), Enterococcus gallinarum (2), Enterococcus casseliflavus(1) , Granulicatella elegans (1), Pediococcus pentosaceus (1) and unidentified isolates (3). Results of Antimicrobial activity of the crude bacteriocins from the isolates against Listeria monocytogenes, Staphylococcus aureus, E. coli and Salmonella sp are shown in the table below. Table 2 above showed the results of the crude bacteriocins against Listeria monocytogenes, Staphylococcus aureus, E. coli and Salmonella sp. Figures 1 - 5 below show the variations in pH of MRS broth inoculated with the isolates and monitored at 24 hourly intervals over a period of 5 days The results of the acid production profile of the isolates are shown in (Figures 1-5). All the isolates produced acid as measured at 5- day intervals.
Table 1: Gram positive cocci bacteria from different sources identified with Vitek 2 compact system.

Table 2: Zones of inhibition produced by cell free extracts of the isolates against the test organisms.
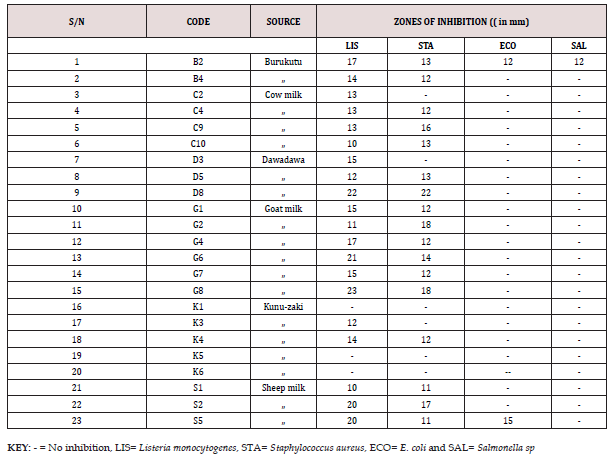
Figure 1: Distribution of isolates according to different hospital fomites.
Keys: WH-Wash Hand basins, TB: Tables, CH: Chairs, DK: Doorknobs, RL: Stair Rails TP: Tap, BD: Beddings.
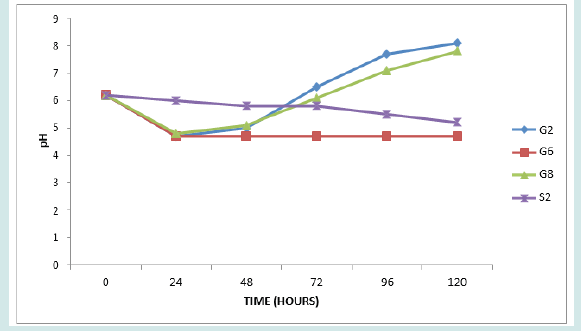
Figure 2: Distribution of isolates according to different hospital fomites.
Keys: WH-Wash Hand basins, TB: Tables, CH: Chairs, DK: Doorknobs, RL: Stair Rails TP: Tap, BD: Beddings.

Figure 3: Distribution of isolates according to different hospital fomites.
Keys: WH-Wash Hand basins, TB: Tables, CH: Chairs, DK: Doorknobs, RL: Stair Rails TP: Tap, BD: Beddings.
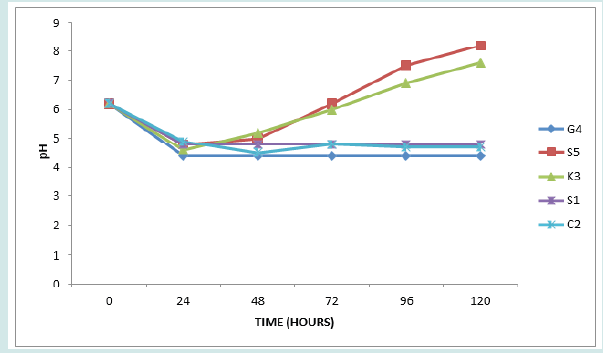
Figure 4: Distribution of isolates according to different hospital fomites.
Keys: WH-Wash Hand basins, TB: Tables, CH: Chairs, DK: Doorknobs, RL: Stair Rails TP: Tap, BD: Beddings.
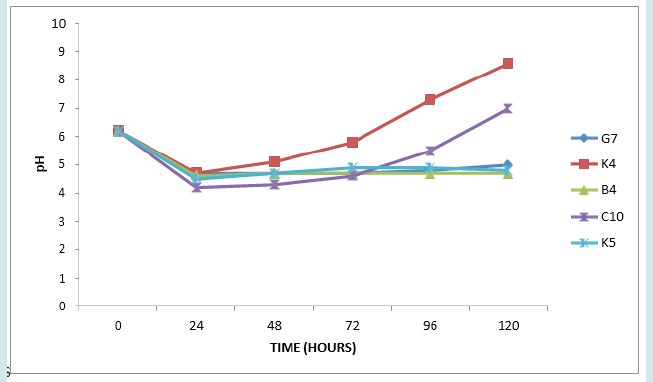
Figure 5: Distribution of isolates according to different hospital fomites.
Keys: WH-Wash Hand basins, TB: Tables, CH: Chairs, DK: Doorknobs, RL: Stair Rails TP: Tap, BD: Beddings.

Discussion
total of eleven different Gram-positive cocci bacteria were identified using Vitek 2 compact system from the six different sources (Table1). The isolates includes Staphylococcus pseudintermedius, Staphylococcus lentus, Staphylococcus sciuri, Kocuria kristinae, Kocuria rosea, Kocuria rhizophila, Enterococcus gallinarum, Enteerococcus faecium, Enterococcus casseliflavus, Pediococcus pentosaceus and Granulicatella elegans. Gram positive cocci bacteria have been isolated from different sources. [16], isolated Gram positive cocci namely Staphylococcus species, Streptococcus species and Lactococcus species from burukutu. [17], also isolated some Gram positive cocci, Streptococci species and Staphylococci species from dawadawa produced from soya bean seeds. [18], isolated Gram positive cocci, Staphylococcus species and Streptococcus species from kunun-zaki. [19], also isolated Gram positive cocci namely Staphylococcus, Streptococcus, Enterococcus, Micrococcus and Lactococcus species from samples of raw milk collected from three areas of high milk production in Zulia state, Venezuela. Similary, [20], isolated Lactococcus, Pediococcus, Leuconostoc, and Streptococcus species from raw milk, cheese and yogurt.
Lactococcus, Enterococcus, Leuconostoc, Pediococcus and Streptococcus species were also identified from raw cow milk [21,22], isolated Enterococcus mundtii, a Gram-positive coccus bacterium from goat milk. Lactic acid bacteria were isolated from all the samples except sheep milk (Table 1). Isolate B4 (Enterococcus casseliflavus), isolate C10 (Enterococcus gallinarum), isolate D3 (Pediococcus pentosaceus), isolate G7 (Enteerococcus faecium,) and isolate K4, K5 and K6 (Enteerococcus faecium, Enterococcus gallinarum and Granulicatella elegans) respectively [23], reported that lactic acid bacteria are ubiquitous bacteria group that are widely spread in nature in niches of diary (fermented), meat and vegetables origin, the gastrointestinal tract and urogenital tract of humans and animals, soil and water. [24], isolated Enterococci species (Enterococcus faecium and E. faecalis.) from a large variety of fresh and prepared foods of animal origin during routine microbiologic control tests in a distribution firm. Similary, [25], isolated Pediococcus pentosaceous from Satar Three isolates, C4, D5 and D8 were unidentified using Vitek 2 compact system (Table 1) apparantly because the organisms are not in the database of the machine.
The catalase test showed that C4 was catalase negative while D5 and D8 were catalase positive. However, microscopic examinations revealed them to be Gram positive cocci. Other methods of identification such as molecular could be explored to get the organisms fully identified. Except for K1, K5 and K6 (Table 2), all the isolates produced substances (bacteriocins) that inhibited the growth of some of the test organisms. This agrees with the report of that the vast majority of all bacterial species appear to be able to produce at least one bacteriocin, most of which are unknown. Generally, Gram positive test organisms were more susceptible to the inhibitory substances (bacteriocins) as each of the substances inhibited either Listeria monocytogenes or Staphylococcus aureus or both. Bacteriocins are known to be proteinaceous or peptidic toxins produced by bacteria to inhibit the growth of similar or closely related bacteria strain(s) [26]. Isolate G8 produced the highest zones of inhibition (23mm) against Listeria monocytogenes and (18mm) against Staphylococcus aureus followed by isolate D8 which produced (22mm) against Listeria monocytogenes and Staphylococcus aureus, followed by G6 with (21mm) against Listeria monocytogenes and (14mm) against Staphylococcus aureus.
Isolate S2 is forth with (20mm) against Listeria monocytogenes and (17mm) against Staphylococcus aureus, S5 with (20mm) against Listeria monocytogenes and (11mm) against Staphylococcus aureus while C9 with (12mm) against Listeria monocytogenes and (16mm) against Staphylococcus aureus and others. The variations in the zones of inhibition by the different cell free extracts on the test organisms could be a reflection of differences in the types and mode of actions of their bacteriocins content (Perez-Ramos et al., 2021) Isolate B2 produced inhibitory substances that has broad spectrum antimicrobial activities as it inhibited both the Gram-positive (Listeria monocytogenes and Staphylococcus aureus) and the Gram negative test organisms (E.coli and Salmonella sp). Isolate S5 also produced antimicrobial substance that has antimicrobial activity against E. coli in addition to Listeria monocytogenes and Staphylococcus aureus. Bacteriocins are known to have a narrow spectrum by inhibiting bacteria taxonomically close or a broad spectrum by inhibiting a wide variety of bacteria [27]. Among the isolates with high antimicrobial activity are Staphylococcus pseudintermedius followed by D8 (partially unidentfied), Kocuria kristinae and Staphylococcus scuri. However, Staphylococcus pseudintermedius is the most frequently occurring isolate [28,29].
Reported that Staphylococcus psudintermedius strain 222 produces a novel peptide bacteriocin termed BacSp222 that inhibited methicillin-resistant S.aureus. Similaly, [30], reported the production of bacteriocin-like substance by Staphylococcus pseudintermedius S28 that inhibited isolates of S. aureus from foods showing inhibition zones of 14.2 to 28.3mm. The unidentified D8 produced antimicrobial substance that has high inhibitory activity next to Staphylococcus pseudintermedius. The lactic acid bacteria isolates: B4, C10, G7 and K4 which are Enterococcus species produced the antimicrobial substances that inhibited either Listeria monocytogenes or Staphylococcus aureus or both. Enterococcus species are known to be one of the most frequent producers of bacteriocins (enterocins), which confer them an advantage to compete in their natural environment (Almeida-Santos et al., 2012). Bacteriocins produced by Enterococci are often similar to those by other lactic acid bacteria (Ness et al.,2014) [31], reported that an isolate named E. faecium LCW 44 from raw camel milk exhibited a broad antibacterial spectrum with an inhibitory activity against several Gram-positive strains belonging to the genera Clostridium, Listeria, Staphylococcus, and Lactobacillus.
Also, Enterococcus casseliflavus M1001 has been shown to produce bacteriocins that have antimicrobial activity against Staphylococcus aureus [32]. Pediococcus pentosaceus (isolate D3) produced an antimicrobial substance that inhibited Listeria monocytogenes with zone of inhibition of 15mm. Pediococcus pentosaceus 147 isolated from artisanal Minas cheese produced with raw milk, and characterized as a bacteriocinogenic strain showed inhibitory activity against different strains of Listeria species (Gutierrez-Cortes et al., 2018). The Kocuria species (isolates C2, G4, K3, S1 and S5) produced antimicrobial substances that inhibited the growth of either one or both of the Gram positive test organisms. Kocuria varians NCC 1482 has been reported to produce a bacteriocin called variacin, which may be applied in food preservation without posing any safety or regulatory issue [33]. The unidentified isolates (D8, D5 and C4) by Vitek 2 compact system, however, produced antimicrobial substances that inhibited the growth of the two Gram positive test organisms. Isolate D8 produced high zones of inhibition (22mm) against both Listeria monocytogenes and Staphylococcus aureus. Isolate D5 has 12mm and 13mm against Listeria monocytogenes and Staphylococcus aureus while C4 produced zones of inhibition of 13mm and 12mm against Listeria monocytogenes and Staphylococcus aureus respectively.
The results of the acid production profile of the isolates are shown in Figures 1-5. All the isolates produced acid as measured in 5 days intervals. However, the acid production varied as shown by the pH readings. Figure 1 presents pH of isolates G2, G6, G8 and S2. These are all members of Staphylococcus psudintermedius. Figure 2 presents pH of isolates G1, B2, K1 and C4. Isolates G1, B2 and C4 are members of Staphylococcus lentus while G1 is Staphylococcus sciuri. [34]. reported that the genera Staphylococci that had an average GC content of 30-39%, produced acid from dextrose and the MRS agar used (TITAN BIOTECH LTD, India) contained 20g/ltr of dextrose thus this agreed with their report. The average GC content of Staphylococcus psudintermedius group genome ranges from 37.4 to 38.3% [35], while the genome sequence of Staphylococcus lentus F1142 has GC content of 31.8% [36]. Figure 3 presents the pH changes of the isolates G4, S5, K3, S1 and C2 which are Kocuria species (Kocuria kristinae, K. rosea and K. rhizophila).
Kocuria species are able to use various carbon sources to produce acids from one or more carbohydrates [36]. Figure 4 presents’ pH changes of the isolates G7, K4, B4, C10 and K5 which are Enterococcus species (Enterococcus faecium, E. casseliflavus and E. gallinarum). Finally, Figure 5 presents changes in pH of isolates D3, K6, C4, D5 and D8, (Pediococcuss pentosaceus, Granulicatella elegans and the three unidentified isolates). Enterococcus species. Pediococcus pentosaceus and Granulicatella elegans are all members of lactic acid bacteria. Lactic acid bacteria are a group of Gram positive, non- spore forming, facultative anaerobes, catalase negative cocci or rods which produce lactic acid as the major end product of fermentation of carbohydrates [37-39]. The changes from initial acidity to alkalinity with the bacterial growth in MRS medium (Figures 1,3 & 4) that correlate with the formation of white precipitates, which increased with increase in alkalinity, is likely due to reactions among the intermediate products of LAB fermentation or the products with the medium component. The nature of precipitate is presently unknown.
Conclusion
Bacteriocin genic Gram positive cocci bacteria are associated with burukutu, cow milk, dawadawa, goat milk, kunu-zaki and sheep milk. The Gram positive cocci bacteria isolated from these samples include genera of Staphylococcus, Kocuria. Enterococcus, Granulicatella and Pediococcus. Their cell free extracts ability to inhibit growth of test organisms with either narrow or broad spectrum activity indicates their bacteriocin producing potential. There is a need to identify, purify and classify the bacteriocins produced by these group of organisms to ascertain their safety and range of activity. The organic acid (s) produced by non-lactic acid bacteria needs to be identified as this may assist in the classification of these genera of bacteria. This research also shows that all the isolates produced acid from dextrose. However, there is apparently no correlation between bacteriocins and acid production.
References
- Yang SC, Lin CH, Sung CT, Fang JY (2014) Antibactreia activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol 5: 241.
- Beninez Chao DF, Leon Buitimea A, Lerma-Escalera JA, MoronesRamirez JR (2021) Bacteriocins: An overview of Antimicrobial, Toxicity and Biosafety Assessment by in vivo Models. Front Microbiol 12: 630695.
- Darbandi A, Asadi A, Ari MM, Ohadi E, Talebi M, et al. (2021) Bacteriocins: Properties and potential use as antimicrobials. J Clin Lab Anal 36(1): e24093.
- Negash AW, Tsehai BA (2020) Current Applications of Bacteriocin. Int J Microbiol 2020: 4374891.
- Papagianni M, Anastasiadou S (2009) Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications. Microb Cell Fact 8: 1-3.
- De Souza Darte AF, Ceotto H, Ceolho MLV, Coelho V, De Paiva Brito MAV, el al. (2013) Identification of new staphylococcins with potential application as food biopreservatives. Food control 3(2): 313-321.
- Almeida Santos AC, Novais C, Peixe L, Freitas AR (2021) Enterococcus spp. as a Producer and Target of Bacteriocins: A Double-Edged Sword in the Antimicrobial Resistance Crisis Context. Antibiotics 10(10): 1215.
- Franz CM AP, Van Belkum M J, Holzapfel WH, Abriouel H, Galvez A (2007) Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev 31( 3): 293-310.
- Jack RW, Tagg JR, Ray B (1995) Bacteriocins of Gram-positive bacteria. Microbiol Rev 59(2): 171-200.
- Riley MA (2009) Bacteriocins, Biology, Ecology, and Evolution. Encyclopedia of Microbiology, 3rd edition.
- Refay RM, Abushady HM, Amer SA, Mailam MA (2020) Determination of bacteriocin- encoding genes of lactic acid bacteria isolated from traditional dairy products of Luxor province, Egypt. Future J Pharm Sci 6(1): 1-10.
- Mohammed SSD, Ijah UJJ (2013) Isolation and screening of lactic acid bacteria from fermented milk products for bacteriocin production. Annals Food Science and Technology 14(1): 122-128.
- Akbar A, Sadiq MB, Ali I, Anwar M, Muhammad N, et al. (2019) Lactococcus lactis subsp. lactis isolated from fermented milk products and its antimicrobial potential. CyTA J Food 17(1): 214-220.
- Schillinger V, Luvke FK (1989) Antimicrobial activity of Lactobacillus’s sake isolated from meat. Applied and Environmental Microbiology. 55(8): 1901-1906
- Remmelsberg M, Radler F (1990) Antimicrobial polypeptides of Lactobacillus species. Journal of Applied Bacteriology 69(2): 177-184.
- Yusuf AB, Gulumbe BH, Kalgo ZM, Aliyu B, Haruna M (2020) Microorganisms Associated with Production of Burukutu ( an alcoholic beverage) in Kebbi state Nigeria. Equity J Sci Technol 7(1): 67-73.
- Tyokusa AG, Iheanetu AN (2018) Bacillus subtilis Isolated from Locally Fermented Soyabean Seeds (Soy-Dawadawa) For Production of Cooking Condiment. Asian J Sci Technol 9(10): 8847-8850.
- Makut MD, Nyam MA, Obiekezie SO, Abubakar AE (2013) Antibiogram of bacteria isolated from kunun-zaki sold in Kefi metropolis. Am J Infect Dis 9(3): 71-76.
- Faria RJ, Garcia UA, Izquierdo CP, Allara CM, Valero LK (2002) Isolation of Gram-positive bacteria from raw milk with antimicrobial residues. Arch Latinoam Nutri 52(1): 68-73.
- Taye Y, Degu T, Fasseha H, Mathewos M (2021) Isolation and Identification of Lactic Acid Bacteria from Cow Milk and Milk Products. The Scientific World Journal 2021: 4697445.
- Wassie M, Wassie T (2016) Isolation and identification of lactic acid bacteria from raw cow milk. Int J Adv Res Biol Sci 3(8): 44-49.
- Laukova A, Fockova V, Simonova MP (2020) Enterococcus mundtii Isolated from Slovak Raw Goat Milk and Its Bacteriocin genic Potential. International journal of Environment Resource Public Health 17(24): 9504.
- Liu W, Pang H, Zhang H, Cai Y (2014) “Biodiversity of lactic acid bacteria,” in Lactic Acid Bacteria, eds H. Zhang and Y. Cai (Dordrecht: Springer) 103-203.
- Devriese LA, Pot B, Van Damme L, Kersters K, Haesebrouck F (1995) Identification of Enterococcus species isolated from foods of animal origin. Int J Food Microbiol 26(2): 187-97.
- Lani NM, Nor NM, Ramli NA, Radhuan Z, Rizan MM, et al. (2015) Effects of Incorporation of Lactic Acid Bacteria on Microbiological Quality and Shelf Life of Raw Satar In book: Beneficial Microorganisms in Food and Nutraceuticals, Microbiology Monographs Edition: 2 Publisher: Spinger International Publishing Switzerland.
- Cotter PD, Ross RP, Hill C (2013) Bacteriocins-a viable alternative to antibiotics. Nat Rev Microbiol 11(2): 95-105.
- Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3(10): 777-788.
- Mills S, Serrano L, Griffin C, Oçonnor PM, Schaad G, et al. (2011) Inhibitory activity of Lactobacillus LMGP-26358 against Listeria innocua when used as an adjunct starter in the manufacture of cheese. Microb Cell Fact 10: S7.
- Wladyka B, Piejko M, Bzowska M, Pieta P, Krzyssik M, et al. (2015) A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci Rep 5: 14569.
- Pinto TS, De Oliveira CP, Da Costa ACV, Lima CO, Barreto HM, et al. (2012) Evidence for production of a bacteriocin-like substance by Staphylococcus pseuintermedius, inhibitory to Staphylococcus aureus from foods. Nat Prod Res 27(12): 1098-1101.
- Vimont A, Fernandez B, Hammami R, Ababsa A, Daba H, et al. (2017) Bacteriocin-Producing Enterococcus faecium LCW 44: A High Potential Probiotic Candidate from Raw Camel Milk. Front Microbiol 8: 865.
- Mikkili I, Venkateswarulu TC, Karlapudi AP, Kodali VP, Srirama K (2019) Characterization of bacteriocin ABC transporter ATP-binding protein produced by a newly isolated Enterococcus casseliflavus M1001 strain. Beni-Suef University of Basic and Applied Sciences 8(1): 1-7.
- Fallico V, McAuliffe O, Ross RP, Fitzgerald GF, Hill C (2011) The potential of lacticin3147, enterocin AS-48, lacticin 481, variacin and sakacin P for food bio preservation. In book Protective cultures, antimicrobial metabolites and bacteriophages for food and beverage bio preservation. Woodhead Publishing Ltd, Cambridge pp, 100-121.
- Auletta AE, Kennedy ER (1966) Deoxyribonucleic Acid Base Composition of Some Members of the Micrococcaceae. J Bacteriol 92(1): 28-34.
- Ben Zakour NL, Beatson SA, Van Den Broek AH, Thoday KL, Fitzgerald JR (2012) Comparative genomics of the Staphylococcus intermedius group of animal pathogens. Front Cell Infect Microbiol 2: 44.
- Nam YD, Chung WH, Seo MJ, Lim SI, Yi SH (1012) Genome sequence of Staphylococcus lentus F1142, a strain isolated from Korean soybean paste. J Bacteriol 194(21): 5987.
- Sahin N, Ay H, Saygin H (2021) Bergey’s Manual of Systematic of Archaea and Bacteria. Willey Online Library.
- Axelson L (1998) Lactic acid bacteria: classification and physiology of lactic acid bacteria. Microbiology and Functional Aspects. Marcel Decker Inc New York.
- Pérez-Ramos A, Madi-Moussa D, Coucheney F, Drider D (2021) Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms 9(10): 2107.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...





