Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2644-1381
Review Article(ISSN: 2644-1381) 
Impact of Similarity Metrics and Dimension Reduction Method on Single-Cell RNA-Sequencing Data Clustering Volume 3 - Issue 5
Yunlin Peng*
- University of Sydney, China
Received: October 10, 2022 Published: November 03, 2022
*Corresponding author: Yunlin peng, University of Sydney, China
DOI: 10.32474/CTBB.2022.03.000173
Abstract
Recently invented high-throughput technique called single-cell RNA-sequencing enables the access of the cell transcriptional profile at the single cell level, which can assist the cell type identification. In order to perform the cell type identification, cell types in the gene expression dataset need to be clustered into several clusters based on similarity metrics between cell types, and each cluster is expected to contain the identical cell type so that the marker genes can be used later for identifying the common cell type for each cluster. Hence, the quality of clustering is essential for ensuring the accuracy of cell type identification. This report aims to verify that if applying the clustering based on similarity matrix of Pearson’s correlation on dataset without PCA will outperform similarity matrix of Euclidean distance on dataset without PCA, Pearson’s correlation on dataset with PCA, and Euclidean distance on dataset with PCA. To achieve this aim, 11 datasets have been applied PCA, and then k-means clustering has been applied on these 11 datasets before and after applying PCA, which results in 4 clustering results for each dataset. The ARI values for each clustering result are calculated to quantify its concordance level with predefined cell-type annotation, and 3 pairwise Wilcoxon rank-sum tests have been performed to verify the statistical significance of the differences among these ARI values. This results that the clustering method of Pearson’s correlation without PCA outperforms the clustering method of Euclidean distance without PCA, while it has similar performance as the clustering methods of Euclidean distance and Pearson’s correlation with PCA.
Introduction
Due to the significant functional consequences of the differences between cells, a method called single-cell RNA-sequencing was recently devel-oped in the field of bioinformatics to assess the cell transcriptional profile at the single cell level [1]. It is useful for assessing the variations amo-ng individual cells in order to identify the cell types, and it is famous for the capability of iden-tifying the mutations in individual cells and the rare cell populations which are lack in the tradit-ional bulk RNA-sequencing [2] Historically, the first publication of single-cell RNA-sequencing was in 2009, and it was applied under the situation of limited biological materials with a small size of single-cell RNA-sequencing libraries which were generated in tubes manually [2,3]. In 2011, the size of the sequencing libraries had been significantly incr-eased with the invention of single-cell tagged reverse transcription sequencing [2]. In 2014, the first commercial platform that made the single cell isolation and library generation a two-step process was available, which resulted in significant time and labor reductions [2]. Soon, another two more efficient library preparation platforms called Drop-seq and Seq-Well were invented in 2015 and 2017, respectively. Spe-cially, the latter one is first portable library prep-aration platform [2]. The framework of single-cell RNA-sequen-cing starts with the dissociation of a group of single cells from a tissue sample. Secondly, a single cell will be isolated from this cell group, and the mRNA will be extracted from this single cell. Next, the mRNA will be converted into cD-NA through the reverse transcription. In order to prepare the sequencing library, an amplified cD-NA will be produced. Finally, a gene expression dataset will be generated after performing the sequencing [1]. Table 1 shows a small subset of a typical gene expression dataset. Though the sizes of different gene expressi-on datasets vary, a common size will be around 50000 rows and 350 columns. Each row in the dataset represents a gene, and each column represents a cell type. All genes within a dataset are unique, while cell types are repetitive. For each dataset, the number of distinct cell types are commonly less than 10. Originally, the values in the dataset are integers, which represent the counts of RNA transcribed from the correspond-ing gene in each cell type with the unit of counts per million [4]. Because of the different total nu-mber of transcriptions in each cell type, which will result in relative bias distance matrix calcul ations in later cell-type clustering process, these values are normalized based on the number of transcriptions in the corresponding cell type [5]. Hence, values in the current dataset are normali-zed versions, representing the expression level of the corresponding gene in each cell type. In order to achieve the goal of cell type identification, these cell types should be cluster-ed into several groups.
Under the ideal situation, all cell types in one group should be identical. Hence, the quality of clustering is essential. However, due to the high dimensionality of the gene expression dataset, clustering datasets with or without dimension reduction might result in different clusters [6]. Moreover, different simila-rity metrics used for clustering might also result in different clusters. Two common similarity metrics are distance-based metrics and correlat-ionbased metrics [4]. Hence, by using the princ-ipal component analysis, Euclidean distance, an-d Pearson’s correlation as representatives of dimension reduction method, distance-based metrics, and correlation-based metrics, respecti-vely, the aim of this report is to discover that during the clustering process, if applying Pearso-n’s correlation on dataset without principal com ponent analysis will outperform applying Euclid-ean distance on dataset without principal compo-nent analysis and applying Pearson’s correlation and Euclidean distance on dataset with principal component analysis. Since previous studies have investigated the impact of different similarity metrics on single-cell RNA-sequencing data clustering and reveal-ed that Pearson’s correlation outperforms other similarity metrics [4], it is worth to investigate that if this result is still tenable after applying principal component analysis on the dataset. Mo-reover, though there are previous studies of impacts of different similarity metrics on clust-ering and the mathematical relationship between principal component analysis and a common clustering algorithm, k-means clustering [6], very few of them discuss the impact of different combinations of similarity metrics and existence of principal component analysis on gene expression data clustering, which will be the main Purpose of this report, and it is worth studied. Methods
Methods
In order to be statistically significant on the results, 11 datasets are analyzed. Each dataset either contains cell types of information from humans or mice.
Data transformation and cleaning
For each dataset, the log2 transformation is applied first to remove outliers and make the dist-ributions of these data approximately normal in order to ensure the later clustering accuracy. Moreover, due to the large amount of data values of 0 in the dataset, these data are increased by 1 before the log2 transformation [4]. Next, a single-cell data specific filtering algorithm called Optimal Gene Filtering for Single-Cell data (OGFSC) is applied on the transformed dataset. Because of the low RNA concentrations from individual cells, these single-cell data are commonly accompanied by extremely high technical noise which should be removed. This algorithm will construct a thresholding curve which is capable to select a subset of genes that can best characterize the data with the minimum size. This goal is achieved by constructing the curve that can best separate the biological noise from the technical noise, which results that the data information, the biological noise, will be well preserved while the level of technical noise is minimized [7].
Principal component analysis
Principal component analysis (PCA) is a common-n unsupervised learning algorithm for dataset’s dimension reduction [5]. In order to test the impact of PCA on cell types clustering, PCA will be applied to each dataset to derive a 10-dimentional dataset from the original large dataset such as dataset with dimension of 50000 x 350. In this case, each row of these datasets will be a principal component, and there are 10 rows in total. Each of these principal components is a linear combination of all original genes. These 10 principal components can account for the vast majority of variations in the dataset. After applying PCA, there are 22 datasets in total, which contain the original 11 datasets and their PCA-version datasets.
Similarity metrics
Pearson’s correlation and Euclidean distance are two commonly used similarity metrics, which are two typical representatives of correlation-based metrics and distance-based metrics. They are used to calculate the distance between two values, which will be used as the judgement for clusterin-g since values with shortest distances will be part-itioned together. These two similarity metrics are calculated as following: Pearson’s correlation coefficient,
𝑑𝑖𝑗=1−Σ(𝑥𝑖𝑔−𝑥̅𝑖)(𝑥𝑗𝑔−𝑥̅𝑗)𝐺𝑔=1√Σ(𝑥𝑖𝑔−𝑥̅𝑖)2𝐺𝑔=1√Σ(𝑥𝑗𝑔− 𝑥̅𝑗)2𝐺𝑔=1;
Euclidean distance,
𝑑𝑖𝑗= √Σ(𝑥𝑖𝑔−𝑥𝑗𝑔)2𝐺𝑔=1,
where 𝑥𝑖𝑔 and 𝑥𝑗𝑔 are the expression level of a gene 𝑔=1,…,𝐺 in cell 𝑖=1,…,𝑁 and cell 𝑗=1,…,𝑁, which are the data values in the dataset, and 𝐺 and 𝑁 are the number of genes and cells in the dataset, which are the number of rows and columns in the dataset, respectively. For a distance matrix 𝐷=(𝑑𝑖𝑗), the element 𝑑𝑖𝑗 represents the distance between cell 𝑖 and cell 𝑗 [4].
K-means clustering
Since these datasets all have known cell-type labels, to assess the impact of different similarity metrics on the clustering performance, for each of these 22 datasets, the k-means clustering with similarity metrics of Pearson’s correlation and Euclidean distance will be applied to partition these cell types into several groups, and these resulted clusters will be compared with the original known cell-type labels. This algorithm starts by randomly selecting k data values, which are the number of distinct cell types in each dataset. These k data values will be treated as the representatives of k clusters, and the distance between the remaining data values to each of these k representatives will be calculated by using the similarity metrics of Pearson’s correlation and Euclidean distance separately, and then the remaining data values will be assigned to their nearest representatives based on the similarity measures. After forming k clusters, the mean of similarity measures within each cluster will be calculated, and the summation of each cluster’s mean similarity measures will be calculated and denoted 𝐽𝑐𝑙𝑢𝑠𝑡. Then each cluste-r’s representatives will be updated to be the current mean value of the cluster. Similarly, all data value will be reassigned to each cluster based on their similarity measures with the new representatives, and 𝐽𝑐𝑙𝑢𝑠𝑡 will be updated. This process will be repeated until this dataset is converged, which means that the value of 𝐽𝑐𝑙𝑢𝑠𝑡 cannot be further minimized [8].
Cluster evaluation measures
After Performing the Clustering, there will be 4 clustering results for each of those 11 datasets, based on different combinations of similarity metrics and the existence of PCA, which are clustering based on Pearson’s correlation, Pearson’s correlation on dataset with PCA, Euclidean distance, and Euclidean distance on dataset with PCA. Each cell type in one cluster is expected to have very similar expression level of genes, and they are expected to be identical cell types. To benchmark the performance of clustering from using these 4 different combinations of clustering methods, a clustering evaluation meas-ure called the adjusted rand index will be applied to quantify the concordance of clustering results on each single-cell RNA-sequencing dataset with respect to their predefined and known cell-type annotations [4]. Firstly, for each clustering result, a confusion matrix shown in Table 2 will be formed to display the concordance of clustering results and predefi-ned cell-type annotations [4]. In Table 2, 𝑎 represents the number of pairs of cell types that are placed in the same class in predefined cell-type and in the same cluster in clustering partition, while 𝑑 represents the number of pairs in different classes and different clusters in both partitions, and 𝑎 and 𝑑 all represent the agreement between the clustering results and predefined cell-type. Conversely, 𝑏 represents the number of pairs of cell types in the same class in predefined cell-type but not in the same cluster in the clustering partition, and 𝑐 represents the reverse. Both of 𝑏 and 𝑐 represent the disagreement between the clustering results and predefined cell-type. Hence, the adjusted rand index (ARI) is used to quantify these agreement and disagreement based on a calcula- tion performed on 𝑎, 𝑏, 𝑐, and 𝑑 as followings [9].
𝐴𝑅𝐼= 2(𝑎𝑑−𝑏𝑐)(𝑎+𝑏)(𝑏+𝑑)+(𝑎+𝑐)(𝑐+𝑑).
Since ARI represents the level of concordan- ce between clustering results and predefined cell-type, it is ranged from 0 to 1, and the closer to 1, the higher the level of concordance, and the better the clustering performance [9]. For each dataset, there will be 4 ARI scores for each different clustering method. In order to compare these 4 ARI values to verify that if applying Pearson’s correlation on dataset without PCA during the clustering process will outperform other 3 methods, 3 pairwise Wilcoxon rank-sum tests will be performed. Wilcoxon rank-sum test is a popular non-parametric test for two independent groups without the assumption of normal distribution of data values within the group [10]. Before performing the test, the ARI values for each dataset will be ranked from 1 to 4, where 1 is for the lowest ARI values and 4 is for the highest ARI values. Next, the ranks for each method in all datasets will be integrated into a list. Hence, four lists containing ranks for four methods will be formed. Since the aim of this report is to verify that if clustering based on Pearson’s correlation without PCA will outperform other 3 methods, then three pairwise Wilcoxon rank-sum tests with the ranks of Pearson’s correlation without PCA against the ranks of other three methods will be performed separately with the alternative hyp-othesis that the rank of Pearson’s correlation without PCA is greater than the rank of other three methods separately.
Results
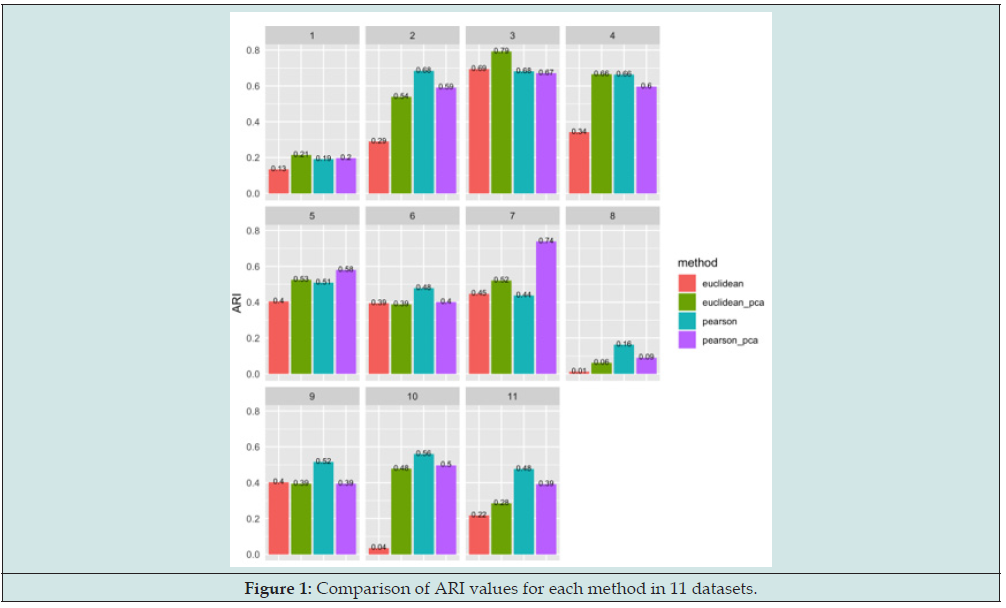
Figure 1 displays the comparison of ARI values for each method in 11 datasets. Each histogram displays the ARI values for one dataset, and values on top of each bar represents its corresp-onding. ARI value. Since the aim is to verify the performance of the clustering method of Pears-on’s correlation without PCA compared to other three clustering methods, then these blue bars in the Figure 1 will be the main target. Based on Figure 1, it is obvious that histograms 2, 6, 8, 9, 10, and 11 displays the successful outcomes since the blue bars in these histograms are higher than other three bars, which represents that clustering using Pearson’s correlation on datasets without PCA results the highest ARI values indicating the best concordance level and clustering perform-ance. Moreover, for histogram 4, there is a tie for the AIR values for clustering method of Pear-son’s correlation without PCA and Euclidean dis-tance with PCA. Conversely, for histograms 1, 3, 5, and 7, the clustering of Pearson’s correlation without PCA does not outp-erform all of other three clustering methods with shorter blue bars and smaller ARI values, which represents the negative outcomes. However, it is remarkable that for most of the histograms, the ARI values for clustering method of Euclidean distance without PCA are all the smallest, which is accord with previous studies that clustering using Euclidean distance does not result in a very favorable performance [4]. In order to verify the statistical significance of results displayed by Figures 1, 3 pairwise Wilcoxon rank-sum tests are performed, whose results are displayed in Table 3.
Table 2: Confusion matrix for measuring the concordance of clustering results with predefined cell type class.
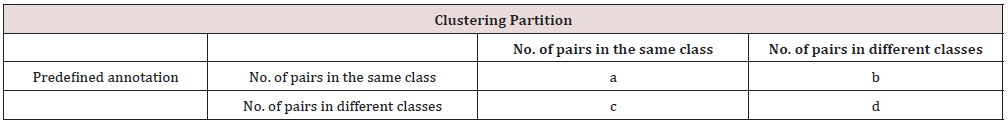
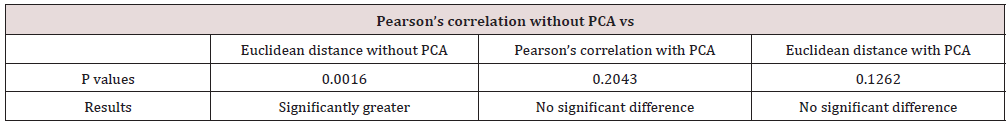
Table 3 displays that the p-value for Wilcox-on rank-sum test with the alternative hypothesis that the rank of ARI values for clustering method of Pearson’s correlation without PCA is greater Than the ranks of clustering method of Euclidean distance without PCA is 0.0016. Under the 5% significance level, this p-value results that the ranks of ARI values of Pearson’s correlation without PCA are significantly greater than the one of Euclidean distance without PCA. This is accord with the result of Figure 1. Conversely, the p-values for other two Wilcoxon rank-sum tests with the alternative hypothesis that the ranks of ARI values for clustering method of Pearson’s correlation with-out PCA is greater that the ranks of ARI values for clustering methods of Pearson’s correlation with PCA and Euclidean distance with PCA are 0.2043 and 0.1262, respectively. Under the 5% significance level, these two p-values results that there is no significant difference between the ranks of ARI values of Pearson’s correlation without PCA and either the one of Pearson’s correlations with PCA or the one of Euclidean distance with PCA. Though the clustering metho-d of Pearson’s correlation without PCA outperfo-rms Pearson’s correlation and Euclidean distance with PCA in 6 out of 11 datasets displayed in Figure 1, the differences between the heights of blue bars and either the green bars (ARI values for Euclidean distance with PCA) or purple bars (ARI values for Pearson’s correlation with PCA) are mostly not prominent. This indicates that Figure 1 is reasonably accord with the results of Wilcoxon rank-sum tests.
Discussion
With respect to the aim of verifying if the cluster-ing method of applying Pearson’s correlation on dataset without PCA will outperform the cluster-ing methods of applying Euclidean distance on dataset without PCA, Pearson’s correlation on dataset with PCA, and Euclidean distance on dataset with PCA, this research can be concluded that the clustering method of Pearson’s correlati-on without PCA outperforms the clustering meth-od of Euclidean distance without PCA, while it has similar performance as the clustering metho-ds of Euclidean distance and Pearson’s correlat-ion with PCA. It is mightbe surprised that performing clustering on datasets after PCA significantly improves the clustering performance when using the clustering method of Euclidean distance, while there is no prominent improvement when using the Pearson’s correlation. Though a previous study has revealed that applying PCA on the dataset can improve its clustering perform-ance [6], there might be a top level of concord-ance performance of each dataset by using k-means clustering due to its self-limitations, since except for the clustering method of Euclidean distance without PCA which has unstable and slightly unfavorable performances, other three methods with stable performances always have similar levels of clustering performances. For instance, in Figure 1, these three methods all result in similar levels of significantly unfavorable performance for datasets 1 and 8, while they all have similarly high levels of performance for datasets 3 and 4. Hence, because of potentially high improvement spaces for clust-ering method of Euclidean distance without PCA, its performance can be improved up to the top level performance of k-means clustering after implementing PCA, which results in the performance of clustering method of Euclidean distance with PCA. However, for the clustering method of Pearson’s correlation without PCA which already reaches the range of the highest performance of k-means clustering proved by the previous study [4], there might not be prominent improvement or even changes on its performanc-es after applying PCA. So, it results that clustering method of Pearson’s correlation without PCA has similar level of clustering performance as clustering methods of Euclidean distance and Pearson’s correlation with PCA. In order to further verify the impact of different combinations of similarity metrics and the existence of PCA on the clustering results, other clustering algorithms such as SIMLR should be implemented, and its results should be compared with the k-means clustering’s.
This strategy can also overcome the limitations of this research which are the onefold implementations of the clustering algorithm and cluster evaluation measure, since these might cause some potential and undetectable biases introduced specifically by k-means clustering and ARI calculation in the research without comparisons with other cluster-ing algorithm and cluster evaluation measures. Hence, applying other cluster evaluation measu-res such as NMI, FM, and Jaccard index might also refine this research [4].
More importantly, this research demons-trates that using the clustering method of Pearson’s correlation or applying clustering on datasets after PCA can result in a relative high clustering performance. After improving and en-suring the accuracy of clusters, it can reduce the difficulty level of using the marker genes to identify the cell type for each cluster since cell types in each cluster are currently highly to be identical, which can assist the cell type identifi-cation [5]. One of the most important purposes of cell type identification is to estimate unknown cell types given their gene expression profiles, which will be achieved by using classification algorith-ms such as k-nearest neighbors. Since k-nearest neighbors algorithm also uses similarity metrics such as Pearson’s correlation and Euclidean distance to calculate similarities between cells, and the quality of classification is also essential, similar approaches can be applied to extend this research to investigate the impact of different si-milarity metrics on classification performances or even the impact of different combinations of similarity metrics used for clustering and classification on the final classification performance.
References
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA (2015) The Technology and Biology of Single-Cell RNA Sequencing. Molecular Cell 58(4): 610-620.
- Wu X, Yang B, Udo-Inyang I, Ji S, Ozog D, et al. (2018) Research Techniques Made Simple: Single- Cell RNA Sequencing and its Applications in Dermatology. The Journal of investigative dermatology 138(5): 1004-1009.
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, et al. (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6(5): 377-382.
- Kim T, Chen IR, Lin Y, Wang AYY, Yang JYH, et al. (2018) Impact of similarity metrics on single-cell RNA-seq data clustering. Briefings in Bioinformatics 00(00): 1-11.
- Hwang B, Lee JH, Bang D (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental & Molecular Medicine 50(8): 1-14
- Ding C, He XF (2004) K-means clustering via principal component analysis. Proceedings, Twenty-First International Conference on Machine Learning ICML 1: 29-38.
- Hao J, Cao W, Huang J, Zou X, Han ZG (2019) Optimal gene filtering for single-cell data (OGFSC)—a gene filtering algorithm for single-cell RNA-seq data. Bioinformatics 35(15): 2602-2609.
- Boyd SP, Vandenberghe L (2018) Introduction to applied linear algebra: vectors, matrices, and least squares. Cambridge University Press, Cambridge, USA.
- Yeung KY, Ruzzo WL (2001) An empirical study on principal component analysis for clustering gene expression data. Bioinformatics 17(9): 763-774.
- Galbraith S, Daniel JA, Vissel B (2010) A study of clustered data and approaches to its analysis. The Journal of Neuroscience 30(32): 10601-10608.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...