Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4579
Mini Review(ISSN: 2637-4579) 
Molecular Simulation Investigation of Prolyl Oligopeptidase from Pyrobaculum Calidifontis and In Silico Docking With Substrates and Inhibitors Volume 2 - Issue 3
Hira Amjad1 ,Waqar Hussain2 andNouman Rasool*3
- 1Department of Chemistry, University of Management and Technology, Lahore, Pakistan
- 2Department of Computer Science, University of Management and Technology, Lahore, Pakistan
- 3Department of Life Sciences, University of Management and Technology, Lahore, Pakistan
Received: May 23, 2018; Published: May 31, 2018
Corresponding author: Nouman Rasool, Department of Life Sciences, University of Management and Technology, Lahore, Pakistan, Email: nouman.rasool@umt.edu.pk
DOI: 10.32474/OAJBEB.2018.02.000141
Abstract
Prolyl oligopeptidase is an important enzyme known to be involved in many human diseases including Alzheimer disease, Parkinson disease and dementia. Microbial world is rich with many diversified enzymes which can be studied as analogous to eukaryotic counter parts. Genome survey of Pyrobaculum calidifontis revealed that three prolyl oligopeptidase with ORFs P_cal 0773; P_cal 1307 and P_cal 1997 are present in this microbe while none of these have been studied. In this study, molecular modeling and in silico characterization of the enzyme prolyl oligopeptidase (P_cal 0773) is performed. The tertiary structure of the enzyme has conserved residues Ser431, Asp513 and His 545 at active site. The structure was observed to have two domains which were α/β hydrolase fold and β-propeller. Where β-propeller domain, most specific feature of Propyl oligopeptidase, is comprised of seven blades constituted by β sheets. The predicted structure was refined using molecular dynamics and docked with two substrates and five inhibitor at catalytic site which showed affinity with the catalytic residues and also forms bond with functional groups of other amino acids present in substrate binding pocket. Among the five inhibitors, three are conventional while two novel inhibitors are also predicted which has different mode of activity for inhibition.
Keywords: Prolyl oligopeptidase, Pyrobaculum calidifontis, β-propeller, Molecular Dynamics, enzymes-substrate complex
Abbrevations: GROMACS: Groningen Machine for Chemical Simulations, OPLS-AA: Optimized Potential for Liquid Simulation, EM: Energy Minimization, NVT: Number Volume and Temperature, NPT: Number Pressure and Temperature, PWE: Particle Mesh Ewald, LINCS: Linear Constraint Solver, GRACE: Graphing Advanced Computation and Exploration, VMD: Visual Molecular Dynamics
Introduction
Prolyl oligopeptidase (POP) represents the family of α/β- hydrolase fold enzymes of serine peptidase. It was first described in 1991, based upon their homology sequences and was identified to have three subfamilies i.e. prolyl oligopeptidase, acylaminoacyl peptidase and dipeptidyl peptidase IV while the forth subfamily i.e. oligopeptidase B was identified latter [1-2]. POP was first discovered as oxytoxin degrading enzyme in human uterus [3]. As it preferentially cleaves the peptides bond on the carboxyl side of proline residues so it was initially named as post-proline enzyme. As proline is an imino acid rather than amino acid hence most of the peptidase are incapable to hydrolyze the peptide bond or the undergo cleavage at proline [4-5]. POP is present in most of the organisms of all three domains of life. It has been isolated and cloned from different organisms like Flavobacterium meningosepticum, Aeromonas hydrophila, Pyrococcus furiosus, Sarcophaga peregrine, Streptomyces morookaensis, Thermococcus sp, Homo sapiens, Trypanosoma brucei, Coprinopsis clastophylla, Escherichia coli, Trypanosoma cruzi, Aspergillus niger, Pleurotus eryngii, Aspergillus oryzaeand Cicer arietinum [6-20]. The homology of these four families are very low but the three dimensional structures are quite similar with each other. The dipeptidyl peptidase and acylaminoacyl peptidase are exopeptidase whereas dipeptidyl peptidase IV is a membrane bounded enzyme and acylaminoacyl peptidase is a cytoplasmic enzyme. PC-POP and oligopeptidase B are endopeptidases as they are present in cytosol. These enzymes do not produce as zymogens but are synthesized as active peptidases. The enzymes or peptidases of this family are mostly large protein molecules (about 80kDa) as compared to other serine proteases and peptidases like trypsin and subtilisin (25- 30kDa). Their most specific property is their selectivity that restrict proteins to 30 amino acids i.e. the peptides undergoing cleavage process must be small thus they do not pose any threat of cleavage to larger molecules present in the cell or nearby the peptidase [21]. The members of this family are involved in many important physiological processes and attract the interest of researchers in pharmaceutics. These are involved in different diseases for example POP is known to play important role in blood pressure control, amnesia and depression, oligopeptidase B in trypanosomiasis. Trypanosomiasis, leishmaniasis and chagas disease are being caused by the prolyl oligopeptidase from trypanosoma cruzi in more than 10 million people worldwide [22]. Actually POP reveals a very high concentration towards brain peptides. The major cause of depression and amnesia is the decrease in POP activity in serum while the lower serum activity also causes bulimia nervosa and anorexia nervosa. It is also considered to be involved in enhancement in Alzheimer disease, may be due to memory loss [23-24].
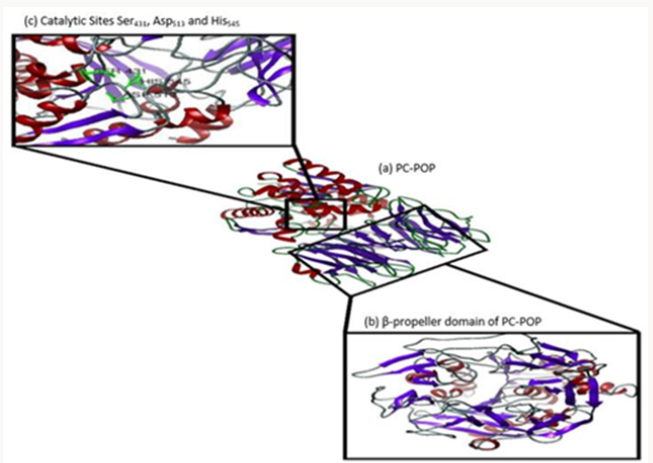
The common structure of POP is composed of a cylindrical shape, consisting of two domains i.e. one is peptidase which is α/β-folds while other is β-propeller domain having β-sheets. The peptidase mostly contains catalytic site of the enzyme [25-27]. The β-propeller domain is based upon seven folds that are the repeats of four stranded anti-parallel β sheets. The N-terminal comprises of β-propeller domain while the α/β folds is C-terminal of the protease. This domain act as substrate filter and gate for allowing the entrance of specific and not large structure [25-29]. As this peptidase belongs to serine protease family hence it has a serine catalytic triad having Serine, His and Asp is present in carboxy terminal region. Which make it is more significant than amino terminal. Pyrobaculum calidifontis is an aerobic facultative obligate hyperthermophilic archaeon [30]. It contains three open reading frames of prolyl oligopeptidase i.e. P_cal 0773, P_cal 1307 and P_cal 1997 while none of these enzymes have been characterized so far. PC-POP with open reading frame P_cal 0773 from P. calidifontis was chosen for the study. Bioinformatics methodology can be utilized as an important prescient apparatus to provide data about structure, function and analysis of these proteins. Molecular dynamics (MD) simulations help in investigating the structure of a biological molecule under some thermodynamic circumstances. These simulations help in studying the protein folding mechanism and structure dynamics analysis [31]. pH of a residue in biological molecule helps in predicting the intra-molecule electrostatic interactions which have a direct influence on the structure of the protein. The most crucial factor for the protein folding is considered to be pH dependent structure stability. The protein folding is the key to many of the biological mechanisms of a biological molecule [32- 33]. Electrostatics represents the charges values along with their positive or negative impacts. It helps in predicting the active site charges and getting to know that whether positive charge ligand will be attracted or negative charge [34]. Hydrophobicity plays an important role in binding and interactions as it increases binding affinity. It also minimizes bond energy as hydrophobicity does not involve water and hydrogen bonds can be formed more freely with the aid of less energy and thus it makes binding more stable [35]. Molecular modelling helps in predicting the structure of an enzyme with the aid of amino acid sequence while the molecular docking techniques help in docking of ligands with the molecules to analyze the inhibitors and substrates activity.
On the basis of characterization of bond length, the strength and type of bond created between protein and ligand can also be determined. The characterization of a bond is based upon the length between hydrogen bond and the donor or the acceptor atom. Thus the classification can be given as the length between 2.2-2.5 Å is said to be the strong covalent bond, 2.5-3.2 Å as moderate but mostly electrostatic bond while the bond length of 3.2-4 Å give rise to a weak electrostatic bond. Most of bond in protein lies in the category of moderate electrostatic bond as the strong covalent bonding requires the specific conditions. It is also a matter to consider that the mostly the hydrogen atoms does not lie in the same straight line when connecting with the donor or the acceptor atom that may result in the elasticity of bond length during the measurement. Thus the mean bond length between hydrogen and the other donor-acceptor atom in protein secondary structure is close to 3.0 Å [36]. In this work, in silico approach was used to provide information about insight of the structural components, sequence, domain arrangement, and catalytic machinery, enzymesubstrate, and enzyme-inhibitor interactions of POP (P_cal 0773) from P. calidifontis.
Materials and Methods
Homology Modelling
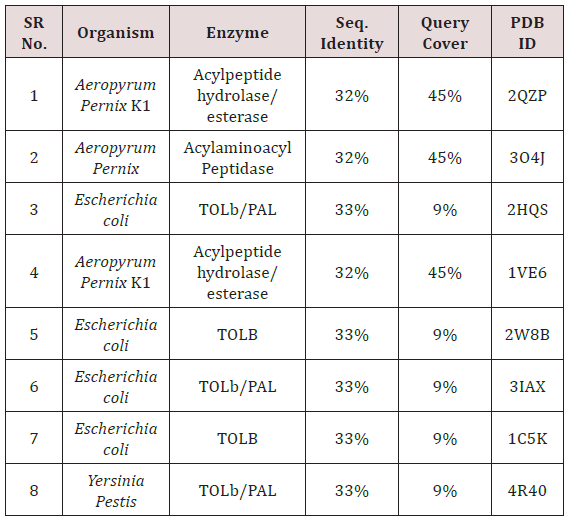
The PC-POP was subjected to homology modeling as no tertiary structure was available. The process included the protein blast on NCBI against RCSB PDB for the quest of suitable templates for modelling. X-ray crystallographic structures having maximum sequence identity in PDB format were chosen as templates. Thus homology modeling was performed using these templates mentioned in Table 1 and the program used for homology modelling was Modeller 9.17 [37].
Molecular Dynamics
Constant temperature MD simulations were performed to study the thermodynamic characteristics and structure dynamics of the PC-POP using Groningen Machine for Chemical Simulations (GROMACS) v 5.0 [38]. Initially, the model of PC-POP was taken as input and all atom force field for optimized potential for liquid simulation (OPLS-AA) was applied to the molecule. MD simulations were performed at 300k temperature along with the constant pH 7 to analyze the thermal stability of the PC-POP. The PC-POP was enclosed in a cubic box by keeping it in the exact center of that box. The distance of the cube walls with the protein at the center was 1.0 Å. Later on, the protein was solvated in water molecules and spc216 water template was used at this phase. At the next phase, the whole system, which is PC-POP solvated in water, was neutralized. The neutralization of PC-POP was performed by the addition of counter ions of Na+ and Cl-. The verlet cut off scheme was used for the neutralization phase and 8 ions were added. After neutralization, the energy minimization (EM) simulation was performed with the aid of the steepest descent algorithm and the maximum number of steps limit for EM was 50000. After minimizing the energy of PC-POP, it was equilibrated with two equilibrations which are constant Number Volume and Temperature (NVT), and constant Number Pressure and Temperature (NPT). Both of the calibrations were performed under 300k and for 100ps. Force fields used for electrostatic potentials in both equilibrations were Particle Mesh Ewald (PWE) with a cubic interpolation whereas hydrogen bonds were adjusted using Linear Constraint Solver (LINCS). Production MD simulation was performed at the end for 1ns. The method was same as equilibrations. The analysis of the results was performed using rms and gyrate utilities of the GROMACS. The graph on the basis of RMSD was plotted for each phase and was viewed using Graphing Advanced Computation and Exploration (GRACE) of data.The molecular dynamics simulations were visualized using Visual Molecular Dynamics (VMD) [39,40].
Electrostatic Potential and Hydrophobicity
Electrostatic potential of the PC-POP was calculated using PDB2PQR [41] facility in Chimera and further Poisson-Boltzmann electrostatics were calculated using ABPS in the chimera [42]. Moreover, the electrostatic potential was also computed using Coulomb’s law and was represented using Coulombic surface coloring in chimera which colors surface according to potential irrespective of explicit hydrogen by generating a grid of potential values. Hydrophobicity analysis of PC-POP was performed using Kyte and Doolittle scale of hydrophobicity and was plotted on the surface of PC-POP using Chimera module of surface coloring on basis of kdHydrophobicity attribute of PDB structure [43].
Molecular docking
The docking of substrates and inhibitor was performed with predicted model in the active pockets of PC-POP. The process was carried out using Autodock and Autodock Vina [44-45]. Two substrates and three out of five inhibitors were known while two of the inhibitors were designed for this study. The substrates, inhibitor and the enzyme were changed into different format by the action of different operations on them in Auto dock. According to the manual, both the protease and substrate model were prepared by adding polar hydrogen bonds and charges in them while grid box was designed to set location surrounding active site and undergo binding [44].
Results and Discussion
Homology modelling
The homology modelling program Modeller 9.17 was used to create the three dimensional model of PC-POP using eight templates with maximum identity with the provided PC-POP sequence (Table 1). Five models created by Modeller 9.17 were analyzed on the basis of their DOPE (Discrete Optimized Protein Energy) values and the one with the lowest dope value of -60514.582031 was considered to be the most suitable model for the further analysis. The DOPE is statistical prospective which is used to evaluate the homology models in predicting protein structure. Ramachandran plot of proposed model also confirmed that over 96 percentages of the residues are in favorable conformation. The structure of PCPOP contains 14 α-helices and 26 β-strands. Among them only 12 α-helices and 7 β-strands are present in its α/β folds while the rest form seven blades in its β-propeller domain. The catalytic residue is Ser431 while the motif around active residue is different for each subfamily. PC-POP has a motif GGSXGGLL, where X is typically Asn or Ala and S is active Ser residue. Among the other residues, Asn381 was the conserved residue among entire serine protease family at residue 456. Asn381 is known to play an important role in the formation of oxyanion hole, which is shown in Figure 1. The Prolyl oligopeptidase from A. pernix form two α/β folds and β-propeller but PC-POP has only one α/β folds and β-propeller while the catalytic site was also found near carboxy terminus. The propeller domain is known to guide the substrate to the catalytic site while acting as a gate for the proper substrate to enter and bind at the catalytic site [46]. POP from trypanosome cruzi (POPTc80) also has the seven blades β-propeller domain along with the same α/β folds [22]. It is reported that it plays an important role in substrate binding [28].
Molecular Dynamics
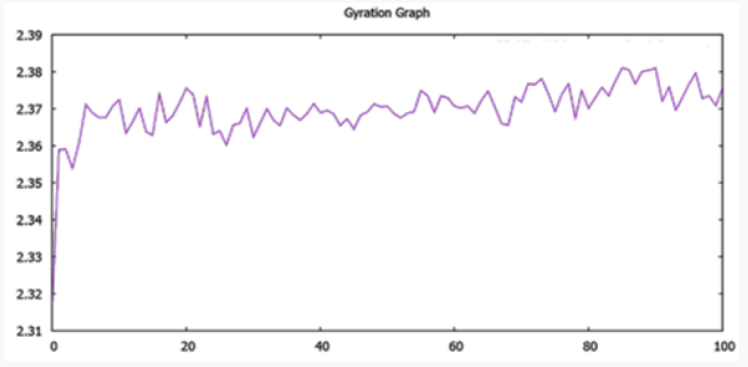
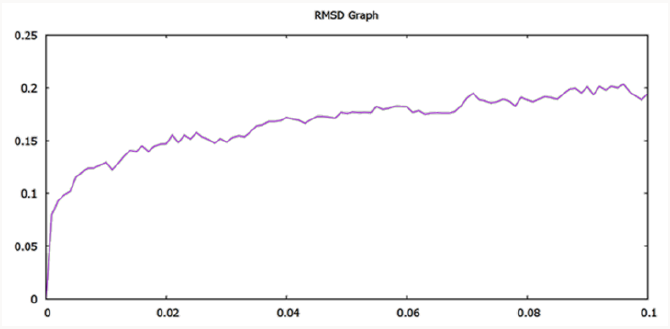
The model was compared with initially predicted model and it gave template match score (TM-score) of 0.966 while root mean square (RMSD) value observed was 1.5Å. As mentioned above that the temperature of the whole simulation along with the equilibrations was set 300k but the RMSD of the structure represents the high stability and rigidity of the POP under thermodynamic circumstances. The radius of gyration (Rg) shows the compactness of the structure and the analysis of Rg shows that the protein is folded under the stable configurations as the value of Rg is steady in the graph which is shown in Figure 2. The model of PC-POP after dynamics, when superimposed with the PC-POP, showed a minor difference in the structure overall, which represents that model is highly stable. The RMSD graph is displayed in Figure 3. Fuxreiter et al. [47] performed the molecular dynamics on POP to study and enhance the behavior of substrate inhibiting, using dynamics simulation techniques. It was observed, with the aid of atomic fluctuations during dynamics simulation, that propeller domain was rigid and did not allow the substrate to interact with active sites. Temperature and pH were kept constant just like in the method proposed here and the POP showed a similarity in stability and rigidity of the structure with the POP in this method [47]. The results were in accordance with Kaszuba et al. [48] as they also reported molecular dynamics of the POP from porcine brain which was bounded with the inhibitor Z-Pro-Prolinal (1QFS PDB) but for simulations and analysis of binding cavity, the inhibitor was removed. The POP from porcine brain was highly stable under thermodynamics circumstances and the structure dynamics showed that there was a minor difference in the structure of POP after the completion of simulation [48]. The dynamics of POP from trypanosoma cruzi (POPTc80) revealed with the aid of RMSD and gyration that the model was compact and highly stable under thermodynamic circumstances of 300k temperature and 1atm pressure [27].
Electrostatic potentials and hydrophobicity
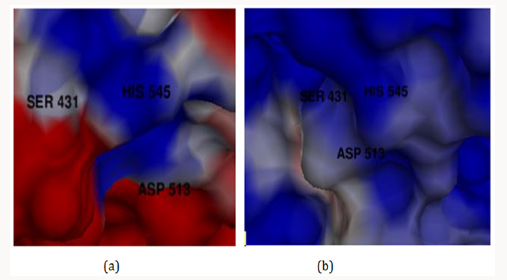
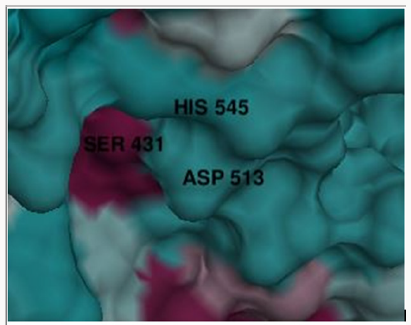
The electrostatic potentials of PC-POP computed are purely positive and would attract the negative charges interactions. The Figure 4 explains the potentials of active sites where the blue color represents positive and red represents negative while white is for neutral. The electrostatic potentials of oligopeptidase B from E.Coli were also studied by constructing a model and then computing molecular potentials. It was observed that active sites have less positive potentials rather than the regular charges which are reported as positive of PC-POP [49]. The analysis of oligopeptidase B from L-amazonesis showed that active sites had less positive electrostatic potentials, same as the enzyme from E.Coli [50]. The hydrophobicity score analysis shows that the binding pocket of PC-POP is highly hydrophobic which represents high interaction rate for hydrophobic amino acids. The hydrophobicity of binding pocket is shown in Figure 5. The color scheme for hydrophobicity is magenta-cyan where cyan represents hydrophobic surrounding and magenta represents hydrophilic surrounding of residue. Hydrophobic interactions are considered as a crucial and important factor for protein folding and the higher hydrophobicity of the POP represents the presence of the additionally packed conformations which would be present due to solvent pressure. In this simulation, it was the spc216 water and at 300k, the higher hydrophobicity represents the higher compactness and decreased solvent pressure [51]. The results show that the hydrophobic substrates and inhibitors would strongly interact with binding site of PC-POP. The SUC-GLY-PRO is purely hydrophobic substrate and its docking study shows the strong hydrophobic interactions, which has also been observed with prolyl oligopeptidase from Trypanosoma Cruzi [27].
Molecular docking
Molecular docking was performed using Autodock and Vina autodock. The purpose of docking was to bind ligands i.e. substrates and inhibitors at the active site of enzyme. The active site consisted of Ser430, Asp513 and His545. While Asn381 is also important for catalytic activity as it is known to form oxy-anion hole for the formation of binding pocket in serine proteases [22].
Docking with substrates
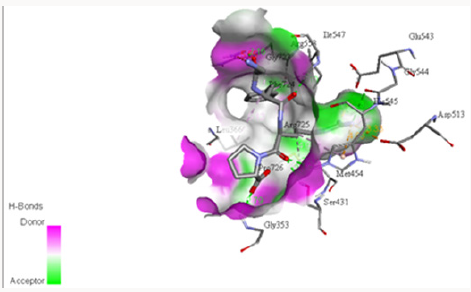
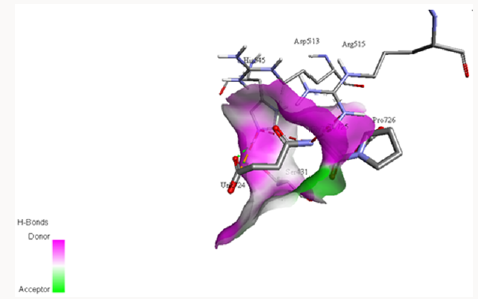
Docking with GLY-PHE-ARG-PRO: The peptidase was docked with GLY-PHE-ARG-PRO ligand which is a tetra peptide, shown in Figure 6. The bond length between the residues was found as His545¬-ligand 2.430Å and 3.953Å, bond length of Ser431-ligand was 2.142Å, bond length of Ile547-ligand was 2.095Å, bond length of Gly430-ligand was 3.070Å and the bond length of Gly433-ligand was 4.6010Å. The binding with catalytic residues Ser431 and His545 was a strong covalent bonding while the interactions with the other residues seems to have electrostatic force of attraction. The binding of GLY-PHE-ARG-PRO ligand with Gly430, Gly433, Ser431 and His545 was observed and binding pattern coincides with that of recombinant POP found in porcine brain. The ARG and GLU of the substrate formed bond with the active residue Ser554 from the catalytic triad of Prolyl oligopeptidase from porcine brain [52]. The same ligand has been reported and docked with the wild type POP bearing pSKPOP and with a mutation of T202C. The binding of GLY-PHE-ARG-PRO ligand with Gly430 and Gly433 was observed while it formed bond with His545 from the catalytic triad [53].
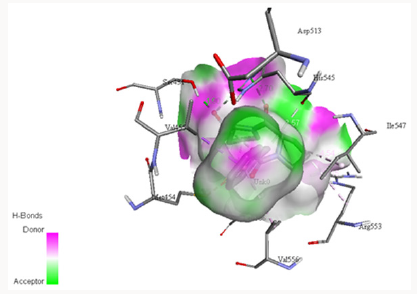
Docking with SUC-GLY-PRO: The PC-POP was also docked with SUC-GLY-PRO ligand as shown in Figure 7. The activity of this substrate have been observed with PC-POP where Gly354 forms two bonds meanwhile bond is made with Ser431 and His545 of the catalytic triad. The presence of small R-group (H) on glycine makes it comfortable to form bond while the bulky groups can cause hindrance for bonds. The bond length of Ser431-ligand was 1.947Å, bond length of Gly354-ligand was 3.862Å while the other bond with same residue had bond length of 3.797Å and the bond length of His545-ligand was 2.175Å. Catalytic Ser431 and His545 formed covalent bonds while the other bonds are resulted by electrostatic force of attraction. The SUC-GLY-PRO ligand has exhibited same binding pattern i.e. His640 and Asp641 with recombinant POP from porcine brain. The presence of Proline residue at the terminal position in substrate causes difference in the residue binding process [54]. The docking of this substrate was first analyzed by Tenorio-Laranga et al. in 2006 with the prolyl oligopeptidase from porcine brain. The binding of this substrate with glycine residue of prolyl oligopeptidase from Wister mice was also observed, which had a specific effect on cognitive skills, specially mood and learning [13]. The same ligand was docked with the prolyl oligopeptidase from Trypanosoma Cruzi. The ligands formed bonds with Arg482 and Arg571. Some other secondary interactions were also found with the Thr475 side chain or Ala477 backbone, along with some favorable hydrophobic interactions [27].
Docking with Inhibitors: The number of inhibitors was docked to figure out its impact on catalytic activity of the enzyme. The binding of conventional POP inhibitors i.e. [2-[[8-(Dimethylamino) octyl]thio]-6-(1-methylethyl)-3-pyridinyl]-2-thienylmethanone oxalate was performed with PC-POP [56]. These inhibitors have a long hydrophobic chain with Thiol group. Hydrophobic chains are known to house at hydrophobic environment of POP and Thiol group forms bond with catalytic residue at active site [56]. Two more inhibitors were designed in this study and their docking was analyzed with PC-POP. The proposed inhibitor were designed by the change in the chemical structure and the formula in order to facilitate the binding of inhibitors at active site of the enzyme. One inhibitor was designed with the addition of a new functional group while in the second, hydrocarbon chain was modified to shorten the length of chain and increasing the inhibition activity.
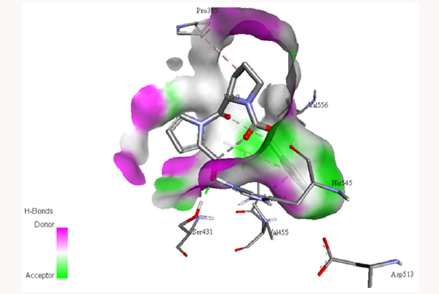
Docking with Z-PRO-PROLINAL inhibitor: Z-PRO-PROLINAL is a known inhibitor for prolyl oligopeptidase [46]. This inhibitor is successfully docked with the PC-POP as shown in Figure 8. The Gly354, Tyr432 and Arg515 along with catalytic triad residues Ser431, Asp513 and His545 of PC-POP formed bonds with Z-PROPROLINAL inhibitor. Gly354, Tyr432 and Arg515 residues are known to play an important role in providing opportunity to inhibitor to get enter to binding site of enzyme. These residues tightly bound with ligand in order to facilitate proteolysis. The bond length of Arg515-ligand is 3.608Å, bond length of Gly354-ligand is 2.46Å while the bond length of Tyr432-ligand is 3.852Å and that with His545-ligand is 3.962Å. Tyr, Gly, Arg of recombinant prolyl oligopeptidase from porcine brain are reported to make bonds with Z-PRO-PROLINAL [48]. Recombinant POP from porcine brain had affinity for Z-PRO-PROLINAL inhibitor at the sites Tyr473 and Arg643 whereas same kind of bonds are formed between PC-POP and Z-PRO-PROLINAL. Z-PRO-PROLINAL inhibitor is involved in the treatment of Alzheimer [55]. Prolyl oligopeptidase enhances the amyloid-β-petpide (Aβ) activity which causes the Alzheimer disease and inhibition of POP reduced the Aβ deposition which cause inhibition in Alzheimer. Also it has been reported that POP inhibitors can exert significant cognition enhancing effects [57]. The same inhibitor has been reported by Puttonen et al. 2006 which inhibits the POP in Homo sapiens. The POP is known to involve in Alzheimer, Parkinson and Huntington disease [58].
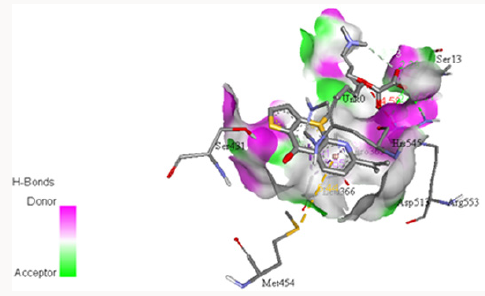
Docking with Pyrrole-2-carboxy-aldehyde: Pyrrole- 2-carboxy-aldehyde, which is a known inhibitor for prolyl oligopeptidase, was docked with PC-POP in order to analyze its binding with catalytic residue for the inhibition of further activity. This potential inhibitor was designed by modification in Z-Pro- Prolinal by adding a pyrrole group instead of pyrrolidine group. Addition of this group is made with a view that it may lead to the stability of due to resonance but the electronegative charge also increase on the aldehydic group with it leading to decrease in its activity. The molecular formula is found to be C18H18N2O4 with molecular mass of 326 g/mol. Thus the bond formed with the catalytic serine after docking with the more positive charge on it. The bonding also formed with Ile547 which is also found to be the active residue. The bond length of Ser431 the oxygen of aldehydic group in inhibitor is 1.829 Å while the bond length of Ile547 with inhibitor is 4.358 Å, as shown in Figure 9. The docking study has shown that Ser431 formed a strong covalent bond with the inhibitor [59]. The similar nature of modification has also been reported earlier [60].
Docking of [2-[[8-(Dimethylamino) Octyl]Thio]-6-(1-Methylethyl)- 3-Pyridinyl]-2-ThienylmethanoneOxalate: PC-POP was docked with the inhibitor Y29794 oxalate, with the molecular formula of C25H36N2O5S2 and molecular mass of 508.694 g/mol. It’s docking with the PC-POP, as shown in Figure 10, revealed that the sulfur and carbonyl groups of inhibitor formed bonds with the catalytic Ser431 and the two active glycines, Gly430 and Gly353. The bond length of Ser431-sulphur is found to be 5.113Å while that of Ser¬¬431¬¬ with oxygen of carbonyl group is 4.395Å. The bond length of Gly430 and sulphur of thiophene is 4.338Å and Gly353 with sulphur of thiophene is 3.73Å. The bonding with the catalytic Ser¬¬431¬¬ and the two active glycine, Gly430 and Gly353 happened to block the binding pocket hence this inhibitor can be recognized as the competitive inhibitor. Thienyl group of inhibitor plays an important role in binding with PC-POP. Thienyl group is actually the sulfur group from thiophene, covering the whole binding site, as reported earlier [22]. Nakajima 1992 firstly reported it as an inhibitor for POP from rat. The inhibitor is known to be effective in the treatment of Schistosomiasis which is induced by POP from the blood fluke of genus Schistosoma in human by blocking the Ser¬¬431 along with the whole active site [63].
Figure 10: Docking of inhibitor [2-[[8-(Dimethylamino) o c t y l ] t h i o ] - 6 - ( 1 - m e t h y l e t h y l ) - 3 - p y r i d i n y l ] - 2 - thienylmethanone oxalate with PC-POP.
Docking with 4‐ {[6‐ (Propan‐2‐Yl) ‐3‐ (Thiophene‐2‐Carbonyl) Pyridin‐2‐Yl] Sulfanyl} Butan‐2‐One: Another inhibitor was designed for this study making two modification in 2-[[8-(Dimethylamino) octyl] thio]-6-(1-methylethyl)-3-pyridinyl]-2-thienylmethanone oxalate i.e. the removal of oxalate group and replacement of diethyl amine with acetate group. The structure was designed using structure sketching utility by PubChem [62]. Later on, it was docked with PC-POP, as shown in Figure 11. The molecular formula for this inhibitor is C17H19NS2O2 with molecular mass of 333g/mol. The inhibitor exhibits great affinity with the active Gly430 and Gly353 which leads in the obstruction in binding of any other substrate at the active site. The bond length between sulphur of thiophene and Gly430 was 4.765Å while the bond length between sulphur of thiophene is 3.079Å. The bond length with both the glycine residues of PC-POP are resulted due to an electrostatic force of attraction existed in them rather any covalent bond. The removal of oxalate makes it a cheaper and less weight molecule can be used for the inhibition process. Thienyl group showed its significance in binding with the PC-POP as reported in earlier studies [64].
Figure 11: Docking of inhibitor 4-{[6-(propan-2-yl)-3- (thiophene-2-carbonyl)pyridin-2-yl]sulfanyl} butan-2-one with PC-POP.
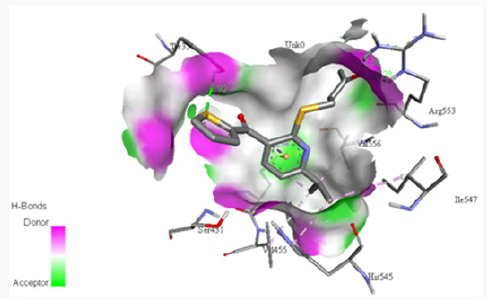
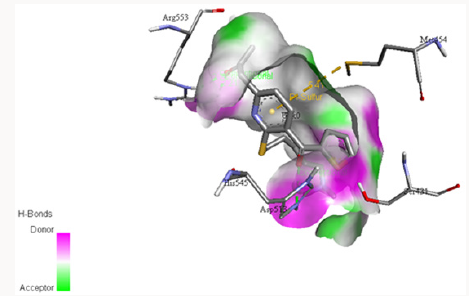
Dockingwith1‐(6‐{[5‐(Dimethylamino)Pentyl]Sulfanyl}‐5‐ (Thiophene‐2‐Carbonyl)Pyridin‐2‐Yl)Propan‐2‐One: Another novel inhibitor was designed with three modifications in [2-[[8-(Dimethylamino)octyl]thio]-6-(1-methylethyl)-3-pyridinyl]- 2-thienylmethanone oxalate i.e. the shortening of hydrocarbon chain, addition of acetate group along and removal of oxalate group, and it was docked with PC-POP. The molecular formula and molecular mass for this molecule is C21H29NS2O2 and 391 g/ mol, respectively. The binding occurs between catalytic Ser431 and Gln358 with the inhibitor, as shown in Figure 12. The bond length between Ser431 and sulphur of thiophene was 3.435Å while that of Ser431 and oxygen of carbonyl was 3.764Å. The bond length of Gln358 and nitrogen of dimethyl amine was 5.11Å. These bond lengths indicate that this bonding is due to the formation of weak electrostatic attraction between the two molecules. The weak attraction between serine and the inhibitor may be attributed to the presence of acetate group with the pyridine which undergoes a with-drawing resonance and inductive effect leading the oxygen of carbonyl to be less electronegative and forming weak bond with more electropositive Ser431 [66]. The binding of catalytic serine residue would result in the inhibition of the enzymatic activity of PC-POP.
Figure 12: Docking of inhibitor 1-(6-{[5-(dimethylamino) pentyl]sulfanyl}-5-(thiophene-2-carbonyl)pyridin-2-yl) propan-2-one with PC-POP.
Conclusion
POP from hyperthermophilic archaea P. calidifontisis was analyzed to examine the activity of enzyme substrate complex and enzyme inhibitor complex using in silico approach. Homology modelling of PC-POP revealed the structure. The structure of PCPOP contains 14 α-helices and 26 β-strands. Among them only 12 α-helices and 7 β-strands are present in its α/β folds while the rest form seven blades in its β-propeller domain. The molecular dynamics, energy minimization and thermodynamic properties to figure out the stability of model meanwhile the molecular dynamics also changed the structure by changing the position of β-sheet blade. Binding site of PC-POP contains hydrophobic region facilitating the substrates containing non-polar amino acids. In order to explore the inhibition of the PC-POP, molecular docking of inhibitors was performed with it. The novel inhibitors are designed on the basis of alteration in functional group from the known inhibitors. The newly designed inhibitors are successfully docked at the active site of the enzyme thus blocking the site for any further binding. Catalytic residues of the binding site forms string bond with these inhibitors. This study explores new dimensions for development of novel and cheap inhibitors against prolyl oligopeptidase which can be used as drugs.
References
- Rawlings ND, Polgar L, Barrett AJ (1991) A new family of serine-type peptidases related to POP. J Biochem 279: 907-908.
- Kanatani A, Masuda T, Shimoda T, Misoka F, Lin XS, et al. (1991) Protease II from Escherichia coli: sequencing and expression of the enzyme gene and characterization of the expressed enzyme. J Biochem 110(3): 315- 320.
- Walter R, Shlank H, Glass JD, Schwartz IL, Kerenyi TD (1971) Leucylglycinamide released from oxytocin by human uterine enzyme. Science 173(3999): 827-829.
- Mentlein R (1988) Proline residues in the maturation and degradation of peptide-hormones and neuropeptides. FEBS Lett 234(2): 2516-2566.
- Cunningham DF, O Connor B (1997) Proline specific peptidases. Biochim Biophys Acta 1343(2): 160-186.
- Chevalier S, Goeltz P, Thibault P, D Banville, J Gagnon (1992) Characterization of POP from Flavobacterium meningosepticum. J Biol Chem 267(12): 8192-8199.
- Kanatani A, Yoshimoto T, Kitazono A, Kokubo T, Tsuru D (1993) Prolyl endopeptidase from Aeromonas hydrophila cloning, sequencing and expression of the enzyme gene: and characterization of the expressed enzyme. J Biochem 113(6): 790-796.
- Harwood VJ, Denson JD, Robinson Bidle KA, Schreier HJ (1997) Overexpression and characterization of a prolyl endopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 179(11): 3613-3618.
- Ohtsuki S, Homma K, Kurata S, Natori S (1997) Molecular cloning of cDNA for Sarcophaga prolyl endopeptidase and characterization of the recombinant enzyme produced by an E. coli expression system. Insect Biochem Mol Biol 27(4): 337-343.
- Szeltner Z, Renner V, Polgar L (2000) Substrate and pH dependent contribution of oxyanion binding site to the catalysis of POP: a paradigm of the serine oligopeptidase family. Protein Sci 9(2): 353-360.
- Nishimura M, Ikeda K, Sugiyama M (2006) Molecular cloning and characterization of gene encoding novel puromycin inactivating enzyme from blasticidin S producing Streptomyces morookaensis. J Biosci Bioeng 101(1): 63-69.
- Lee HS, Kim YJ, Cho Y, Kim SJ, Lee JH (2007) Characterization of POP from hyperthermophilic archaeon Thermococcus sp NA1. J Biosci Bioeng 103(3): 221-228.
- Tenorio Laranga J, Venalainen JI, Mannisto PT, Garcia Horsman JA (2008) Characterization of membrane bound prolyl endopeptidase from brain. FEBS J 275(17): 4415-4427.
- Bastos IM, Motta FN, Charneau S, Santana JM, Dubost L, et al. (2010) POP of Trypanosoma brucei hydrolyzes native collagen: peptide hormones and is active in the plasma of infected mice. Microbes Infect 12(6): 457- 466.
- Chen JT, Chao ML, Wen CY, Chu WS (2012) Screening, purification and characterization of an extracellular POP from Coprinopsis clastophylla. J Microbiol 50(4): 652-659.
- Motta FN, Bastos IM, Faudry E, Ebel C, Lima MM, et al. (2012) The Trypanosoma cruzi virulence factor oligopeptidase B (OPBTc) assembles into an active and stable dimer. PLoS One 7(1): 30431.
- Kang C, Yu XW, Xu Y (2013) Gene cloning and enzymatic characterization of an endoprotease Endo Pro Aspergillus niger. J Ind Microbiol Biotechnol 40(8): 855-864.
- Arima J, Tokai S, Chiba M, Ichiyanagi T, Yabuta Y, et al. (2014) Gene cloning and biochemical characterization of eryngase: a serine aminopeptidase of Pleurotus eryngii belonging to the family S9 peptidases. Biosci Biotechnol Biochem 78(11): 1856-1863.
- Kang C, Yu XW, Xu Y (2014) Purification and characterization of a prolyl endopeptidase isolated from Aspergillus oryza. J Ind Microbiol Biotechnol 41(1): 49-55.
- Sharma R, Suresh CG (2015) Genome wide identification and structure function studies of proteases and protease inhibitors in Cicer arietinum (chickpea). Comput Biol Med 56: 67-81.
- Polgar L (2002) The POP family. Cell Mol. Life Sci 59: 349-362.
- MD Bastos I, N Motta F, Grellier P, Santana JM (2013) Parasite prolyl oligopeptidases and the challenge of designing chemotherapeuticals for Chagas disease, leishmaniasis and African trypanosomiasis. Current medicinal chemistry 20(25): 3103-3115.
- Maes M, Goossens F, Scharpe S, Calabrese J, Desnyder R, et al. (1995) Alteration in plasma POP activity in depression: mania: and schizophrenia: effects of antidepressants: mood stabilizers and antipsychotic drugs. Psychiatry Res 58(3): 217-225.
- Maes M, Monteleone P, Bencivenga R, Goossens F, Maj M, et al. (2001) Lower serum activity of POP in anorexia and bulimia nervosa. Psych neuroendocrinology 26: 17-26.
- Kiss AL, Szeltner Z, Fulop V, Polgar L (2004) His507 of acylaminoacyl peptidase stabilizes the active site conformation: not the catalytic intermediate. FEBS Lett 571(1-3): 17-20.
- Bastos IM, Grellier P, Martins NF, Cadavid Restrepo G, De Souza Ault MR, et al. (2005) Molecular, functional and structural properties of the prolyl oligopeptidase of Trypanosoma cruzi (POP Tc80), which is required for parasite entry into mammalian cells. Biochemical Journal 388(1):29-38.
- De Almeida H, Leroux V, Motta FN, Grellier P, Maigret B, et al. (2016) Identification of novel Trypanosoma cruzi prolyl oligopeptidase inhibitors by structure based virtual screening. Journal of Computer Aided Molecular Design 30(12): 1165-1174.
- Fulop V, Bocskei Z, Polgar L (1998) POP: an unusual b-propeller domain regulates proteolysis. Cell 94(2): 161-170.
- Fulop V, Szeltner Z, Polgar L (2000) Catalysis of serine oligopeptidase is controlled by a gating filter mechanism. EMBO Reports 1(3): 277-281.
- Amo T, Paje MLF, Inagaki A, Ezaki S, Atomi H, et al. (2002) Pyrobaculum calidifontis sp nov: a novel hyperthermophilic archaeon which grows under atmospheric air. Archaea 1(2): 113-121.
- Anwer K, Sonani R, Madamwar D (2015) Role of N-terminal residues on folding and stability of C-phycoerythrin: simulation and urea-induced denaturation studies. Journal of Biomolecular Structure and Dynamics 33(1):121-33.
- Flanagan MA, Ackers GK, Matthew JB (1981) Electrostatic contributions to the energetics of dimer-tetramer assembly in human hemoglobin: pH dependence and effect of specifically bound chloride ions. Biochemistry 20(26): 7439-7449.
- Song Y, Mao J, Gunner MR (2009) MCCE2: improving protein pKa calculations with extensive side chain rotamer sampling. Journal of computational chemistry 30(14): 2231-2247.
- Tsuchiya Y, Kinoshita K, Nakamura H (2004) Structure‐based prediction of DNA‐binding sites on proteins using the empirical preference of electrostatic potential and the shape of molecular surfaces. PROTEINS: structure, Function, and Bioinformatics 55(4): 885-894.
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Structure, Function, and Bioinformatics 11(4): 281-296.
- Jeffrey GA (1997) An introduction to hydrogen bonding. Oxford University Press, UK.
- Webb B, Sali A (2016) Comparative Protein Structure Modeling Using Modeller. Current Protocols in Bioinformatics.
- Abraham MJ, Murtola T, Schulz R (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software X 30(1): 19-25.
- Turner PJ (2005) XMGRACE, Version 5.1. 19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton.
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. Journal of molecular graphics 14(1): 33-38.
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic acids research 32: 665-667.https://www.ncbi.nlm.nih.gov/pubmed/?term=Baker%20NA%5BAuthor%5D&cauthor=true&cauthor_uid=15215472
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. (2004) UCSF Chimera-a visualization system for exploratory research and analysis. Journal of computational chemistry 25(13): 1605-1612.
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. Journal of molecular biology 157(1):105-132.
- Morris GM, Ruth H, Lindstrom W, Sanner MF, Belew RK, et al. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility: J Comp. Chem 30(16): 2785-2791.
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function: efficient optimization and multithreading. J Comp Chem 31(2): 455-461.
- Fulop V, Szeltner Z, Renner V, Polgar L (2001) Structures of POP Substrate/Inhibitor Complexes: Use of inhibitor binding for titration of the catalytic histidine residue. J Biol Chem 276(2): 1262-1266.
- Fuxreiter M, Magyar C, Juhasz T, Polgár L, Simon I (2005) Flexibility of POP: molecular dynamics and molecular framework analysis of the potential substrate pathways. Proteins: Structure, Function, and Bioinformatics 60(3): 504-512.
- Kaszuba K, Rog T, St. Pierre JF, Mannisto PT, Karttunen M, et al. (2009) Molecular dynamics study of POP with inhibitor in binding cavity. SAR and QSAR in Environmental Research 20(7-8): 595-609.
- Gerczei T, Keseru GM, Naray-Szabo G (2000) Construction of a 3D model of oligopeptidase B, a potential processing enzyme in prokaryotes. Journal of Molecular Graphics and Modelling 18(1): 7-17.
- de Matos Guedes HL, Carneiro MP, Gomes DC, Rossi-Bergmanmn B, Giovanni de Simone S (2007) Oligopeptidase B from L. amazonensis: molecular cloning, gene expression analysis and molecular model. Parasitology research 101(4): 853-863.
- Szeltner Z, Rea D, Renner V, Fulop V, Polgar L (2002) Electrostatic Effects and Binding Determinants in the Catalysis of POP: Site Specific Mutagenesis at the Oxyanion Binding Site. J.Biol.Chem 277(45): 42613- 42622.
- Szeltner Z, Juhasz T, Szamosi I, Rea D, Fülöp V, et al. (2013) The loops facing the active site of POP are crucial components in substrate gating and specificity. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1834(1): 98-111.
- Szeltner Z, Rea D, Renner V, Juliano L, Fülop V, et al. (2003) Electrostatic environment at the active site of POP is highly influential during substrate binding. J Biol Chem 278(49): 48786-48793.
- Peltonen I, Mannisto PT (2011) Effects of diverse psychopharmacological substances on the activity of brain prolyl oligopeptidase. Basic & clinical pharmacology & toxicology 108(1): 46-54.
- Szeltner Z, Rea D, Juhasz T (2002) Substrate-Dependent Competency of the Catalytic Triad of POP. J.Biol.Chem 277: 44597-44605.
- Charalambous A, Mangner TJ, Kilbourn MR (1994) Synthesis of (N‐ [11C] methyl) Y‐29794, a competitive inhibitor of prolyl endopeptidase. Journal of Labelled Compounds and Radiopharmaceuticals 34(6): 499- 504.
- Rossner S, Schulz I, Zeitschel U (2005) Brain prolyl endopeptidase expression in aging, APP transgenic mice and Alzheimer’s disease. Neurochemical research 30(6-7): 695-702.
- Puttonen KA, Lehtonen S, Raasmaja A (2006) A prolyl oligopeptidase inhibitor, Z-Pro-Prolinal, inhibits glyceraldehyde-3-phosphate dehydrogenase translocation and production of reactive oxygen species in CV1-P cells exposed to 6-hydroxydopamine. Toxicology in vitro 20(8): 1446-1454.
- Berg, Jeremy M, John L (2007) Biochemistry, WH Freeman and Company, NewYork.
- Palko M, Molnar Z, Kivela H (2009) Ring closure reactions of bicyclic prolinol and prolin ester enantiomers. Arkivoc (6): 221-234.
- Birney YA, O Connor BF (2001) Purification and characterization of a Z-Pro-prolinal-insensitive Z-Gly-Pro-7-amino-4-methyl coumarinhydrolyzing peptidase from bovine serum-a new proline-specific peptidase. Protein expression and purification 22(2): 286-298.
- Racys DT, Rea D, Fulop V, Vills M (2010) Inhibition of POP with a synthetic unnatural dipeptide. Bioorganic & medicinal chemistry 18(13): 4775- 4782.
- Fajtova P, Stefanic S, Hradilek M, Dvořák J, Vondrášek J, et al. (2015) POP from the blood fluke Schistosoma mansoni: from functional analysis to anti-schistosomal inhibitors. PLoS Negl Trop Dis 9(6): e0003827.
- Jarho EM, Venalainen JI, Poutiainen S, Leskinen H, Vepsäläinen J, et al. (2007) 2 (S)-(Cycloalk-1-enecarbonyl)-1-(4-phenyl-butanoyl) pyrrolidines and 2 (S)-(aroyl)-1-(4-phenylbutanoyl) pyrrolidines as POP inhibitors. Bioorganic & medicinal chemistry 15(5): 2024-2031.
- Ihlenfeldt WD, Bolton EE, Bryant SH (2009) The PubChem chemical structure sketcher. Journal of cheminformatics 1(1): 1.
- DeRuiter J (2005) Resonance and induction tutorial. Principles of drug action I. Auburn University, Alabama 1-1.
Editorial Manager:
Email:
biomedicalengineering@lupinepublishers.com

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...