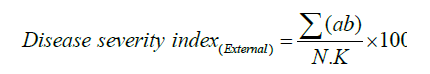Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4676
Research Article(ISSN: 2637-4676) 
Induction of Systemic Acquired Resistance in Papaya by Foliar Application of HrpN Recombinant Protein for Increased Resistance against Papaya Dieback Pathogen Volume 2 - Issue 3
Norliza Abu Bakar1*, Mohd Zulfadli Sohaime1, Nor Mustaiqazah Juri1, Rafidah Badrun1 and Johari Sarip2
- 1Biotechnology and Nanotechnology Research Centre, Malaysian Agricultural Research and Development Institute, Malaysia
- 2Horticulture Research Centre, Malaysian Agricultural Research and Development Institute, Malaysia
Received: April 18, 2018; Published: April 24, 2018
Corresponding author: Norliza AB, Biotechnology and Nanotechnology Research Centre, Malaysian Agricultural Research and Development Institute, Malaysia
DOI: 10.32474/CIACR.2018.02.000136
Abstract
Plants are continuously exposed to undesirable pest and pathogen threats. In response, plants have developed numerous mechanisms to protect themselves against the pathogens. Systemic acquired resistance (SAR) is an inducible disease resistance response in plant species. It is found in a large range of plant species including papaya and characterized by broad spectrum disease control and an associated coordinated expression of a set of pathogenesis related (PR) genes and proteins which are also known as SAR markers. Expression and purification of HrpN from Erwinia mallotivora, the causal agent of papaya dieback, was carried out. In this report, HrpN recombinant protein was tested and characterized for its effect and potential as elicitor that can increase papaya defence against E. mallotivora through the activation of SAR mechanism. Based on disease severity analysis, control plants which were untreated, showed faster disease infection rate and severity when compared to the recombinant protein treated plants. Increased resistance towards the papaya dieback pathogen was shown to be associated with increased expression of selected plant defined genes using quantitative Real Time analysis which were observed after the papaya was sprayed with the recombinant HrpN protein. Based on physiological and molecular analysis, the selected protein has induced SAR; increased selected SAR associated defence gene expression and increased the papaya resistance against the papaya dieback pathogen.
Keywords: Systemic acquired resistance; Recombinant protein; Erwinia mallotivora
Abbreviations: SAR: Systemic Acquired Resistance; E. mallotivora: Erwinia mallotivora; SA: Salicylic Acid; Hrp: Hairpin Proteins; PR: Pathogenesis-Related; MTI: MAMP-triggered immunity; ETI: effector-triggered immunity; SA: salicylic acid; BTH: Benzo Thiodiazole
Core Ideas
a) HrpN recombinant protein was tested and characterized for its effect and potential as elicitor that can increase papaya defence against E. mallotivora through the activation of SAR mechanism.
b) Increased selected SAR associated defence gene expression and increased papaya resistance against the papaya dieback pathogen observed.
Introduction
Papaya is a popular and commercially available fruit in the tropical and subtropical regions. It is highly known not only for its nutritious quality but also for its medicinal functions [1]. During its prime time, Eksotika, Sekaki and Solo were the Malaysia’s flagship varieties for export with an export value of about RM100–120 million per year, a total volume of 58,149 mt which accounted for 21% of the global trade in 2004 [2,3]. Papaya dieback disease caused by Erwinia mallotivora is the main cause for the rapid decline of Malaysian papaya production, amounting to 60% decrease in papaya production [4,5]. When attacked by pathogen, plants defend themselves through activation of plant defence mechanism which includes oxidative burst of cells, alteration in cell wall composition and de-novo synthesis of compounds like phytoalexin and elevated expression of pathogenesis-related (PR) proteins. The plant defence mechanisms include MAMP-triggered immunity (MTI), effectortriggered immunity (ETI) and systemic acquired resistance (SAR) signify different layers of active plant defence strategy [6]. Plants also have the ability to activate quantitative protection against extensive spectrum of microorganisms upon inoculation with a pathogen, exogenous application of proteins from microorganism or through application of chemicals [7].
The elevated resistance of the whole plant is known as SAR which is an inducible defence response present in a wide range of plant species including papaya [8]. Systemic plant resistance or Systemic acquired resistance involves a salicylic acid (SA)-mediated pathway of defence reactions within the plant [9,10]. During activation of SAR, induced plants showed an earlier boost of exogenous salicylic acid and activation of pathogenesis- related (PR) protein genes [11,12]. Production of PR genes/proteins can lead to increased resistance against pathogen attack [13,14]. Initiation and activation of PR proteins cascade can be produced by exposing the plant to a virulent, avirulent, and nonpathogenic microbe, or molecules with low molecular weight and sometimes volatile molecules such as salicylic acid and jasmonate [15- 17]. The inductions of SAR by using external inducers have been investigated in the past in plants such as tobacco [18] and Arabidopsis thaliana [19]. Incitation of defence reaction occurs not just at the establishment of pathogen recognition but additionally in distal regions of the plant and can last for weeks upon induction [20]. SAR is an effective innate immune response that offers protection against certain infection of pathogens. SAR may also be introduced by treating plants with salicylic acid (SA) and SA analogues; 2,6- dichloroisonicotinic acidity (INA) and benzothiodiazole (BTH) [21-23].
Phytopathogens are known to secrete proteins and virulence factors collectively known as effectors that are essential for pathogenesis and colonization of their host plants [24]. Pathogenicity of E. mallotivora depends on these effectors, which control the pathogen ability to cause disease and to elicit specific defence responses in papaya plants [25]. Erwinia mallotivora genome was already sequenced and bioinformatics tools were utilised to predict genes that potentially encode virulence factors and toxins along with other molecules that promote pathogenesis [26]. Like many other plant pathogenic bacteria, E. mallotivora contains type III secretion system (T3SS) that delivers effectors proteins into host plant. The T3SS apparatus is a key virulence determinant in many Gram-negative plant bacteria. Due to its importance, a lot of studies have been conducted to disable or block the function of T3SS by targeting known T3SS processes [27]. This could serve as a method to control plant microbial-associated diseases.
Effector proteins as virulence factors are known to suppress diverse signalling pathways required for plant innate immunity [28]. Apart from being effectors, some effectors proteins, known as hairpins, have been revealed to elicit plant defence and SAR responses [29]. Type III secreted hairpins are glycine-rich and also heat-stable proteins that are secreted from Gram-negative plantpathogenic bacteria. The hairpin proteins have been proven to elicit defence response and activate SAR for increased disease tolerance against diverse plant pathogens [23]. In selected cases, during fungial, oomycetal or plant pathogen attack, increased defence responses without the symptom exhibited by hypersensitive response cell death were recorded in plants treated with foliar application of hairpins proteins or genetically modified plants that constitutively expressed hairpins genes. This was observed in Arabidopsis after spray treatment with E. amylovora HrpN [30]. Activation of activated SAR in the plant conferred disease resistance to Hyaloperonospora sp. and Pseudomonas syringae pv. Tomato and in addition stimulated the expression of the pathogenesis-related (PR) 1 genes [31].
Another successful research finding includes reduced diseases caused by Phytophthora infestans and Botrytis cinerea in tomato through application of HrpN hairpin proteins [32]. Rice sprayed with Hpa1, another type of Hrp protein also showed strong resistance to X. oryzae pv. oryzae and Magnaporthe grisea [33]. Past studies have implied SAR strategy as another useful approach for controlling plant diseases through the activation of host plant defences by application of various agents or external inducers. Thus, this research aims to assess application of selected recombinant hairpin protein from the papaya dieback pathogen for SAR activation as an alternative new strategy to control papaya dieback disease. In an effort to develop recombinant proteins as potential SAR inducer, cloning and expression of HrpN from E mallotivora was carried out in a bacterial system. Our study is carried out to evaluate the effectiveness of HrpN recombinant protein in inducing Systemic Acquired Resistance (SAR) in papaya for enhanced disease resistance to papaya dieback pathogen.
Materials and Methods
Bacterial strains and growth conditions
Escherichia coli strains, Top10 (Invitrogen,USA) and BL21, were cultivated and grown in LB medium at 37°C respectively. Antibiotic ampicillin (Amp) was used at the concentration of 50μg/ml where required. Erwinia mallotivora was grown in LB broth at 28 °C.
Recombinant protein cloning, expression and purification
The HrpN gene was isolated from Erwinia mallotovora. Sets of primers, termed HrpN forward (ATGAGTCTGAATACGAGTCC) and reverse (GCCGCGTCAGTTTGCTTCGT) was designed to incorporate selected restriction enzymes sites to facilitate the cloning processes. Erwinia mallotivora DNA was isolated from E. mallotivora grown in LB broth at 28°C overnight using bacterial genomic extraction kit (Sigma Aldrich). Genomic extractions were carried out according to the manufacturer’s instruction. The polymerase chain reaction (PCR) was performed using the E. mallotivora DNA as the template and specific primers that target the HrpN region. Cycle parameters for PCR included an initial incubation time of 3 min at 95°C, 30 cycles of 30 sec at 94°C, 1 min at 55°C for annealing and 1 min at 72°C for extension, and followed by final elongation for 10 min at 72°C. The PCR products were visualised by agarose gel electrophoresis, gel-purified and cloned into pGEMT (Promega) according to the manufacturer’s instruction. Transformed cells were plated out on the LB plate supplemented with 100g/ml ampicillin and 20g/ ml X-gal to allow blue and white colonies selection. The gene was then subjected to sub clone into Pet-20b expression vector and transformed into BL21 E. coli expression strain. For expression of HrpN, the PET-20b/HrpN transformed bacteria were selected on LB agar plates containing 100g/ml ampicillin. A single colony of the transformed bacteria was inoculated in 5.0ml LB medium containing appropriate antibiotic for overnight cultivation at 37°C.
Aliquots of the culture were inoculated into 50ml LB medium with 50g/ml ampicillin at 37°C until the OD600 reached 0.5. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM to induce the expression. The expression was carried out for six hours at 37°C and the bacteria were harvested afterwards by centrifugation at 2500g for 15 minutes. The bacterial pellet was resuspended in 20mM Tris-HCl, pH 8.0, and lysed with a sonicator or treated with Bugbuster reagent (Novogen). For confirmation analysis, lysate, soluble and insoluble fractions (pellet) from each expression were analysed by SDSPAGE and Western Blotting using specific anti-His antibody. For large scale purification, the expressed HrpN cells were treated with Bugbuster reagent (Novogen) to be lysed then purified using Ni-NTA column (His tag protein purification) via the Acta Prime Chromatography System. The purified proteins were quantified using Bradford protein assay and also analyzed by SDS PAGE followed by Coomassie Brilliant Blue Staining. For Western Blot analysis, 20μg of protein from each samples were separated by 10% SDS PAGE and transferred to PVDF membrane before being probed using anti-His antibody with gentle agitation for 2h and further incubated for 2h with anti-mouse IgG alkaline phosphatase conjugate. The membrane was washed, added with the substrate solution (BCIP/NBT) and incubated until the bands appeared.
Plant Growth
Carica papaya (Eksotika I) seeds were germinated in small polystyrene cups containing potting soil. In addition to these, two months old papaya seedlings were obtained from MARDI Pontian. Johor, Malaysia. Soil rich in organic matter and nutrients with a mix of compost was used. Fertilisation and watering were conducted accordingly. The plants were continued to be grown in the greenhouse at the MARDI glasshouse house complex under glasshouse conditions.
Recombinant protein application and pathogen inoculation
A set of formulation treatments and controls were tested for their effectiveness in inducing SAR and protecting the plants against papaya dieback disease. Protein inducer treatments were carried out in 4-6 months old papaya seedling using foliar spray application solution for each seedling for three times at one week interval. Each seedling was inoculated (at the first three nodes) with 10ml of pathogen (E. mallotivora) at the concentration of 1x106 cfu one week after the third inducer treatments. Water treated plants were included as control.
Symptom evaluation
After treatments with recombinant protein and the pathogen inoculation, effects of the pathogen inoculation were evaluated through disease severity (DS) statistical analysis. After treatments with salicylic acid (SA), effects of the pathogen inoculation were evaluated for disease severity. For disease severity, index 5 (on 1 to 5 scales) on each plant was recorded according to stem blackening and the mean value was calculated. For evaluation of stem blackening, they were recorded according to the scale of 0=symptomless, 1=leaf vein blackening, 2=leaf vein blackening and slightly wilting, 3=leaf stalk wilting, 4=stem blackening and 5=plant died. Data was analysed using analysis of variance (ANOVA) followed by comparison of means using Duncan multiple range test (DMRT) [34].
Tissue collection, RNA extraction and pathogenesis-related gene analysis via RT qPCR
For molecular analysis, leaves were collected on day 20 after the first recombinant HrpN foliar application, frozen in liquid nitrogen, and stored at -80°C until further analysis. Leaf tissue was grounded to a fine powder, and RNA was extracted using GeneJet (Thermo Scientific) kit following the manufacturer’s instruction. For qPCR, 2ug of RNA was DNAse- treated to remove genomic DNA contamination and the transcripts were converted into cDNA using Biorad Reverse Transcription in accordance with the manufacturer’s protocol. The resulting cDNA was used as the template for qPCR using primers designed based on known pathogenesis-related proteins in papaya [35]. Two housekeeping genes-actin and 40SRNP-were used as the reference genes for normalization of the expression fold. SensiFast SYBR Hi-ROX kit (Bioline, USA) was used for the RT-qPCR following the manufacturer’s protocol. The experiment was carried out using Bio-RAD CFX96 real-time PCR system (Bio-Rad,USA ). The expression profiling graph was plotted using the Bio-RAD CFX96 Manager software (Bio-Rad, USA).
Results and Discussion
Cloning, expression and purification of selected recombinant protein
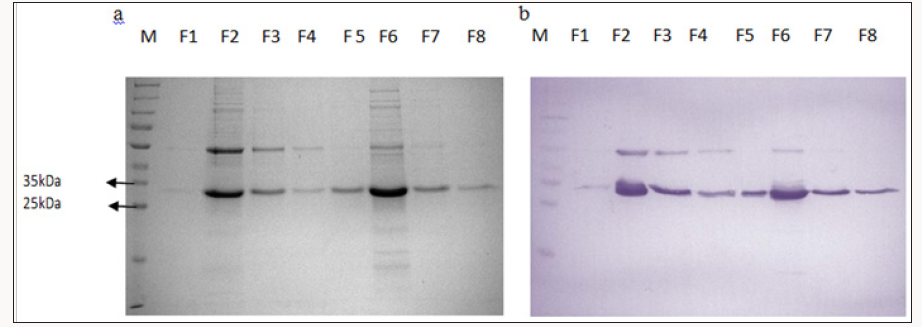
In this report, we attempted to look for the after-effect of foliar spraying of a T3SS protein termed HrpN from E. mallotivora, the causal agent of papaya dieback disease in Malaysia. The likelihood of activation of disease resistance mechanism via SAR against the pathogen in papaya was investigated. Formerly, Peng [36] showed that hpa1 gene of Xanthomonus oryzae pv. oryzae enhanced defence responses to diverse pathogens in tobacco. For this study, cloning of HrpN from E. mallotivora was carried out to produce recombinant HrpN. In an effort to develop a bacterial expression system for selected HrpN from E. mallotivora, gene encoding HrpN was inserted into the pET-20b(+) bacterial expression vector from Novagen. Based on the restriction enzyme digestion and sequencing analysis, we have successfully cloned HrpN gene into pET-20b(+) expression vector. Expression of the HrpN gene in pET-20b expression vector was induced in the presence of isopropyl-beta- D-thiogalactopyranoside (IPTG). The proteins of the uninduced lysates, induced lysates, the soluble fraction and the insoluble fraction were analyzed using SDS–PAGE, stained with Coomassie Blue and Western Blot analysis. The molecular weight of the expressed proteins was equivalent to the predicted molecular weight of the HrpN which was estimated to be approximately 30kDa.
Recombinant HrpN proteins were expressed as fusion proteins with an N-terminal His tag, enabling affinity purification of proteins using Nickel NTA column. To obtain larger amount of expressed HrpN, the expression was carried out in large scale (2 litre cultures) and purified 35kDa 25kDa using the Ni-NTA affinity column via Acta Prime Chromatography System. After purification, the major contaminating bands and impurities were eliminated during the affinity chromatography process. 20ug of proteins from each fraction were analyzed using SDS–PAGE, stained with Coomassie Blue and Western Blot using the anti-His antibody. Figure 1 shows the chromatogram of elution fractions of HrpN using Ni-NTA column. The fractions containing the intended HrpN recombinant proteins were either freeze-dried or stored at -80ºC for further usage. Approximately 50-100mg of pure recombinant protein was normally obtained from every 2 litre cultures of LBBroth expression culture, induced with 0.5mM IPTG, 37°C for 2 hours. The amount of pure HrpN protein was sufficient for the foliar application treatment of papaya seedlings to determine the effect of SAR inducement for increased tolerance to papaya dieback disease.
Figure 1: SDS PAGE (a) and Western Blot (b) analysis of recombinant HrpN proteins obtained through large scale expression after purification via inclusion bodies prep and Ni- NTA affinity column.(M: Protein ladder, F1-F8 : Fractions collected after Ni-NTA affinity column).
Recombinant protein treatments and pathogen inoculation/ pathogen infection assay for SAR assessment
The HrpN recombinant protein was tested to evaluate its effectiveness in inducing SAR in papaya for elevated disease resistance response and to suppress the development of papaya dieback disease. The experiment consisted of 10 replicates for each treatment and control, and was conducted at MARDI’s Biotechnology & Nanotechnology infection house using 4 monthold papaya seedlings arranged in Randomized Completely Block Design (RCBD) with two control treatments. Tap water was used to water the plants daily. Standard fertilisation and pest control programs were applied for plant maintenance. Protein inducer treatments were carried out using foliar spray application solution for each seedling for three times at one week interval. Water treated plants were included as control. The effect of HrpN protein treatment on plant vigour was assessed beforehand for two months. However, no differences in plant height, stem diameter and root mass were observed between control and HrpN- treated plants indicating there was no effect of the treatments on the plant health. To assess the protein ability to increase papaya tolerance against the papaya dieback pathogen, inoculation of ~1x 108 E. mallativora was carried out on the treated seedlings three week after the first foliar spraying for the response to disease symptoms and inducer treatments. Disease development was supervised based on quantitative assessment by assessing percentage of Disease Severity (%DS). Disease severity treatment were computed based on the formulation below using the disease symptoms scoring of 0=symptomless, 1=leaf vein blackening, 2=leaf vein blackening and slightly wilting, 3=leaf stalk wilting, 4=stem blackening and 5=plant died. The disease severity index (DSI) was computed according to the formula described by Campbell and Madden [37] and Kim [34].
Where,
DSI=Disease Severity Index
Σab=Sum of the product of assessed plants with their corresponding score scale
N=Total number of assessed plants K=Highest score scale.
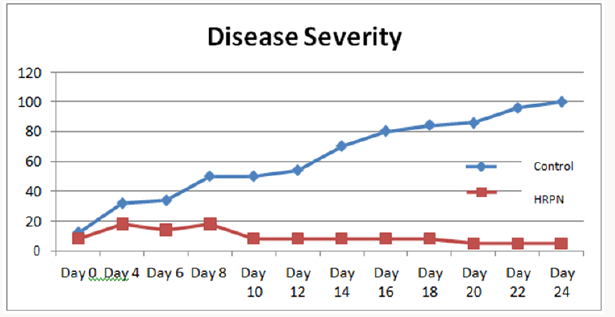
Three to four days after pathogen challenge, papaya dieback disease symptom was visually rated by assessing the percentage of disease progress for the disease severity assay until 25 days post infection. In general, the HrpN formulation showed a reduced degree of symptom compared to the water control in two repeated trials. Results presented here demonstrated that the formulation increased disease tolerance to papaya dieback as previously demonstrated. The disease severity assay (DS) measure was used to indicate the effectiveness of treatments in suppressing the disease. The disease symptom was shown to develop much slower in the seedlings treated papaya plants compared to positive control treatment. Both treated and control plants started showing the stage 1 symptoms of papaya dieback disease approximately on day 4. However, the disease severity percentages were observed more on control plants when compared to treated plants. Subsequently, the severities of disease in control plants increased rapidly with 70% severity on day 14 and continue to rise until day 24 where all controls were observed to succumb to the pathogen infection. Interestingly reverse effect was observed in treated plants. Although plants in both groups exhibited the stage 1 symptom approximately on the same day post infection, the disease severities were shown to decrease in treated plants post infection with the bacterial dieback pathogen. Although initial disease symptom of brown discoloration was observed at early days post infection, all of the leaves in treated plants that showed the early symptoms started to drop between day 6 to day 10 post infection, and new shoot continued to be produced. These resulted in the decreased of severity in treated plants as shown in Figure 2.
The analysis of the disease severity demonstrated that the treated papaya plants showed significantly lower disease severity compared with positive control treatments. Based on statistical analysis, there was highly significant relationship between control and the HrpN-treated plants. The obtained result clearly revealed that the HrpN was effective in increasing resistance against papaya dieback pathogen. The use of hairpin proteins from pathogen has been shown to increase the host against the intended pathogen. Choi [38] reported enhanced disease resistance to both X. oryzae pv. oryzae and Magnaporthe grisea in rice and Arabidopsis plants that were highly expressed with hpa1 gene. An elicitor, pemG1, a hairpin gene which was isolated from M. grisea was also shown to increase disease resistance in transgenic rice containing the hairpin gene. The expression of defence related phenylalanine ammonia-lyase genes were also observed [8]. Similarly, the HrpN formulation showed promising results in inducing SAR in papaya after the papaya seedlings were applied with the protein. The development of the disease symptoms was much slower when compared to positive control treatment. The result suggests that application of SAR inducers certainly has the potential to suppress the development of papaya dieback disease.
Real Time qPCR validation analysis
Analysis of defence mechanism can provide valuable details for papaya dieback disease management strategies. This will offer valuable information for the development of durable, economical, and broad spectrum management approach for the disease. Accordingly, to determine the effect of the formulations on the expression of selected papaya defence genes expression, leaves from papayas that were applied with formulations and control plants were taken from each treatment and control replicates. Each sample has at least three biological replicates representing different individual trees. All samples were ground and stored in -80°C freezer. RNA extraction method was carried out using Plant RNA extraction kit (Thermo Scientific) following the manufacturer’s instruction. Through this method, RNAs extracted were shown to be intact and had a high concentration (Figure 3). The RNAs obtained were transcribed and used for Real Time PCR analysis. Previously, Norliza [35] showed that several pathogenesis related genes which include PR-1b, PR 1, PR1d and NPR1 have the potential to be used as SAR markers due to the increasing levels of genes expression levels few days post treatment with known SAR inducers. These genes were used to investigate the plant defence response after application with the inducers.
Figure 3: RNA obtained from treated and control plants for validation with SAR markers via Real Time PCR. C1-C3 are control untreated plants while T1-T3 are plants treated with HrpN recombinant proteins.
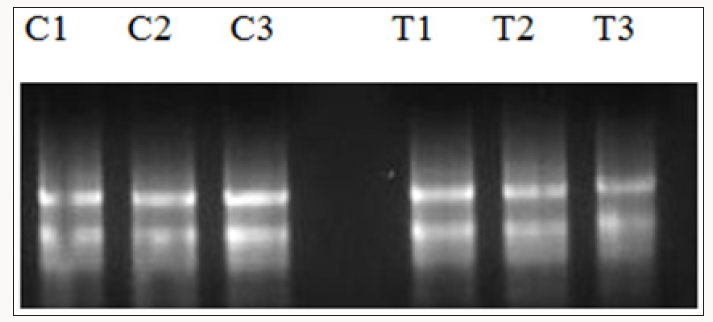
Figure 4: Normalized fold expression of PR1d and NPR1 in control plant (non-treated) and recombinant HrpN protein treated papaya leaf tissues.
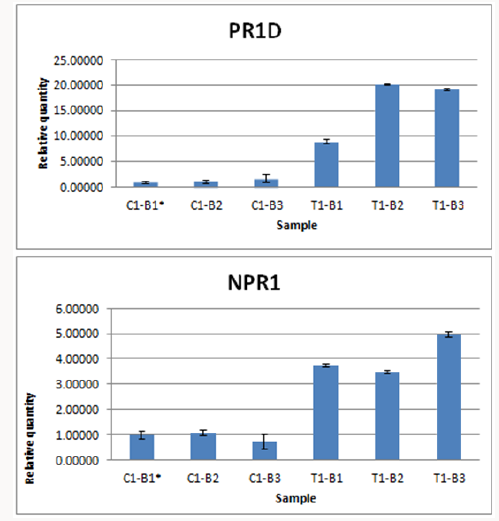
The expression profile of two defence genes, PR1d and NPR1 that were correlated with SAR inducements in papaya in control plant (non-treated) and recombinant HrpN protein treated papaya leaves tissues was conducted with Actin and 40snp used as the reference gene for normalisation [39,40]. As shown in Figure 4, the normalised fold expression of recombinant HrpN treated papaya leaves tissues were higher than the expression in control plant tissues. By using ΔΔCT method, the increased in expression fold of ~10 to ~20 of PR1d gene in papaya plants treated with HrpN recombinant proteins in comparison of each control were observed. Seemingly, the fold increased expression of NPR1 gene was also observed in plants treated with HrpN recombinant proteins in comparison of each control with fold changes around 3-5 fold (Figure 4). Salicylic acid (SA) is a vital hormone in plant immunity and NPR1 is a gene that is triggered by SA. NPR1 is known to be involved in the SAR activation for the regulation of plant defence genes. NPR1 has been shown to induce pathogenesis related (PR) proteins after pathogen attack such as by bacteria and fungi [41]. NPR1 which contains conserved ankyrin repeat domain, a broad complex, tramtrack, and bric-à-brac/poxvirus and zinc finger (BTB/POZ) domain is the master regulator of salicylic acid mediated responses. In Arabidopsis, NPR1 was shown to control the beginning of SAR and other immune signaling pathways for basal defence and mediating crosstalk between SA along with other phytohormones. During genetic screens for mutants defective in SA responses; mutants with defects in NPR1 failed to resolve various SAR-inducing treatments, displaying little expression of pathogenesis related (PR) genes and exhibiting elevated the likelihood of infections [42,43].
Interestingly, NPR1 shares similar structural features with mammalian immune cofactor IκB, that engages in crucial roles in inflammation, immunity, cell proliferation, differentiation, and survival [44]. Norliza [40] showed that a set of PR defence related proteins were not significantly expressed in E. mallotivora infected plants through iTRAQ and quantitative Real Time PCR. These data indicated that the expression of the selected PR genes were not high enough to protect the papaya from the pathogen attack. However, upon application of recombinant HrpN, the expression fold of PR1d genes was increased to ~10 to ~20 in three different treated plants. SAR marker gene pathogenesis-related gene 1 (PR1) which was isolated from Brassica juncea and named as BjPR1 also demonstrated elevated expression in leaves of B. juncea after Alternaria brassicae infection via Quantitative real-time PCR (qRT-PCR) analysis. Furthermore, BjPR1 gene was shown to be strongly induced following SA treatments, suggesting its roles in SAR mediated plant defence [45]. From the quantitative Real Time PCR analysis, it was suggested that the application of recombinant HrpN increased the plant defence related gene expression that are related to the SAR. Furthermore, the expression pattern of the selected genes has the potential to be used in the development of molecular markers for the identification of resistant cultivars or donor varieties for molecular breeding of papaya for increased tolerance or resistance against the papaya dieback pathogen [46].
Conclusion
Erwinia mallotivora HrpN was successfully cloned and expressed in the E. coli system. Foliar application of the HrpN recombinant protein was tested to evaluate its effectiveness in inducing SAR in papaya for enhanced disease resistance to papaya dieback pathogen. Phenotypic data was taken to see if there was any effect of the recombinant protein to the papaya plants. It was concluded that recombinant protein is safe to be used as SAR chemical inducer. Control plants, which were untreated, showed faster disease infection rate when compared to treated plants as shown by the disease severity assay. It can be concluded that for positive SAR inducement, recombinant HrpN is sufficient to enhance the defence system of papaya to combat papaya dieback disease.
Acknowledgment
Evans EA, Ballen, FH (2012) An overview of global papaya production, trade, and consumption. Topics: Food and Resource Economics, Extension service Institute of Food and Agricultural (IFAS): 1-7.
References
- Evans EA, Ballen, FH (2012) An overview of global papaya production, trade, and consumption. Topics: Food and Resource Economics, Extension service Institute of Food and Agricultural (IFAS): 1-7.
- Rabu MR, Mat Lin R (2005) Prospect of papaya in the world market: Malaysia perspective. In Proceeding of First International Symposium on Papaya, Genting Higlands, Malaysia, 22-24.
- Anon (2009) Patient died of papaya. Dead disease.
- Noriha MA, Hamidun B, Rohaiza AR, Indu Bala SJ (2011) Erwinia mallotivora sp., a new phatogen of papaya (Carica papaya) in Peninsular Malaysia. International Journal of Molecular Sciences 12(1): 39-45.
- Chan YK, Baharuddin AG (2010) Rejuvenating the flagging papaya industry in Malaysia: the role of MAFC. Acta Hort (ISHS) 851: 37-40.
- Shunping Y, Xinnian D (2014) Perception of the plant immune signal salicylic acid. Current Opinion in Plant Biology 20: 64-68.
- Yan Y, Tang L, Hu J, Wang J, Adelakun TA, et al. (2018) Munronin O, a potential activator for plant resistance. Pesticide Biochemistry and Physiology 146: 13-18.
- Qiu XH, Guan P, Wang ML, Moore PH, Zhu YJ, et al. (2004) Identification and expression analysis of BTH induced genes in papaya. Physiological and Molecular Plant Pathology 65(1): 21-30.
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3(10): 1085-1094.
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185-209.
- Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Ann Rev Phytopath 44: 135- 162.
- Chaturvedi R, Shah J (2007) Salycilic acid in plant disease resistance. In salycilic acid-A Plant Hormne. S Hayat, A Ahmad Springer pp. 335-370
- Zhu T, Song F, Zheng Z (2006) Molecular characterization of the rice pathogenesis-related protein, OsPR-4b,and its antifungal activity against Rhizoctonia solani. J Phytopathol 154: 378-384
- Borad V, Sriram S (2008) Pathogenesis-Related Proteins for the Plant Protection. Asian J Exp Sci 22(3): 189-196.
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266(5188): 1247-1250.
- Xu Y, Chang, PL, Liu D, Narasimhan ML, Raghothama KG, et al. (1994) Plant Defense Genes Are Synergistically lnduced by Ethylene and Methyl Jasmonate. Plant Cell 6: 1077-1085.
- Wu C, Bradford KJ (2003) Class I Chitinase and â-1,3-Glucanase are differentially regulated by wounding, methyl jasmonate, ethylene and gibberellin in tomato seeds and leaves. Plant Physiol 133(1): 263-273.
- Sticher L, Mauch Mani B, Métraux JP (1997) Systemic acquired resistance. Annu Rev Phytopathol 35: 235-270.
- Mauch F, Mauch Mani B, Gaille C, Kull B, Haas D, et al. (2001) Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J 25(1):67-77.
- Burns EE, Keith BK, Refai MY, Bothner B, Dyer WE (2018) Constitutive redox and phosphoproteome changes in multiple herbicide resistant Avena fatua L. are similar to those of systemic acquired resistance and systemic acquired acclimation. J Plant Physiol 220: 105-114.
- Kamatham S, Pallu R, Pasupulati AK, Singh SS, Gudipalli P (2017) Benzoylsalicylic acid derivatives as defense activators in tobacco and Arabidopsis. Phytochemistry 143: 160-169.
- Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact 19(2): 123-129.
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839-863.
- Prasannath K (2013) Pathogenicity and Virulence Factors of Phytobacteria. Acad J Biosci 1(1): 24-33.
- Norliza AB, Rafidah B, Lina R, Laziana A, Rohaiza AR, et al. Identification and validation of putative Erwinia mallotivora effectors via quantitative proteomics and Real Time Analysis. J Agric Food Tech 7(9): 1.
- Rohaiza AR, Bakar NA, Rozano L, Badrun R, Amin NM, et al. (2014) Draft genome sequence of Erwinia mallotivora BT-MARDI, causative agent of papaya dieback disease. Genome Announcements 2(3): e00375-14
- Degrave A, Fagard M, Perino C, Brisset MN, Gaubert S, et al. (2008) Erwinia amylovora type three-secreted proteins trigger cell death and defense responses in Arabidopsis thaliana. Mol Plant Microbe Interact 21(8): 1076-1086.
- Wu HJ, Wang AH, Jennings MP (2008) Discovery of virulence factors of pathogenic bacteria. Current Opinion in Chemical Biology 12(1): 1-9.
- Anastasia PT, Skandalis N, Anastasia D, GaziMarina NB, Sarris Panagiotis F, et al. (2010) Playing the “Harp”: evolution of our understanding of hrp/hrc genes. Annual Review of Phytopathology 48: 347- 370.
- Degrave A, Moreau M, Launay A, Barny MA, Brisset MN, et al. (2013) The bacterial effector DspA/E is toxic in Arabidopsis thaliana and is required for multiplication and survival of fire blight pathogen. Mol Plant Pathol 14: 506-517.
- Liu R, Lü B, Wang X, Zhang C, Zhang S, et al. (2010) Thirty-seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. Journal of Biosciences 35(3): 435-450.
- Fontanilla M, Montes M, De Prado R (2005) Effects of the foliarapplied protein “Harpin(Ea)” (messenger) on tomatoes infected with Phytophthora infestans. Commun Agric Appl Biol Sci 70(3): 41-45.
- Chen L, Zhang SJ, Zhang SS, Qu S, Ren X, et al. (2008) A fragment of the Xanthomonas oryzae pv. oryzicola harpin HpaG Xooc reduces disease and increases yield of rice in extensive grower plantings. Phytopathology 98(7): 792-802.
- Kim HS, Hartman GL, Manandhar JB, Graef GL, Steadman JR, et al. (2000) Reaction of soybean cultivars to sclerotinia stems rot in field greenhouse and laboratory evaluations. Crop Science 40: 665-669.
- Norliza AB, Rafidah B, Mohd Zulfadlie S (2016) Characterization of Pathogenesis Related (PR) Genes expression associated with disease resistance for the establishment of Systemic Acquired Resistance Marker. Plant Genomic Congress Asia. Kuala Lumpur, Malaysia, pp. 11-12.
- Peng JL, Bao ZL, Ren HY, Wang JS, Dong HS (2004) Expression of harpin(xoo) in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phytopathology 94(10): 1048- 1055.
- Campbell CL, Madden LV (1990) Introduction to Plant Disease Epidemiology. John Wiley & Sons, New York, USA.
- Choi MS, Heu S, Chon PN, Jong KH, Sook LJ, et al. (2012) Expression of HPA1 gene encoding a bacterial harpin protein in Xanthomonas oryzae pv. oryzae enhances disease resistance to both fungal and bacterial pathogens in rice and Arabidopsis. The Plant Pathology Journal 28: 364- 372.
- Wee CY, Muhammad Hanam H, Mohd Waznul Adly MZ, Khairun HN (2014) Expression of defense-related genes in papaya seedling infected with Erwinia mallotivora using real-time PCR. Journal of Tropical Agriculture and Food Science 42(1): 73-82.
- Norliza AB, Nor Mahfuzah Husna S, Rozeita L, Rafidah B (2015) iTRAQ proteins analysis of early infected papaya plants with papaya dieback pathogen. ASIAN Journal of Plant Biochemistry 2(1): 10-14.
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, et al. (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1(6): 639-647.
- Canet JV, Dobon A, Roig A, Tornero P (2010) Structure-function analysis of npr1 alleles in Arabidopsis reveals a role for its paralogs in the perception of salicylic acid. Plant Cell Environ 33: 1911-1919.
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88(1): 57-63.
- Oeckinghaus A, Ghosh S (2009) The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1(4): a000034
- Ali S, Mir ZA, Bhat JA, Tyagi A, Chandrashekar N, et al. (2018) Isolation and characterization of systemic acquired resistance marker gene PR1 and its promoter from Brassica juncea. Biotech 8(1): 1027-1028.
- Ollitrault P, Bruyere S, Ocampo JA, Lapeyre L, De Gallard A, et al. (2005) Papaya breeding for tolerance to bacterial decline (Erwinia sp.) in the Caribbean region. Acta Horticulturae 1: 80-92.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...