Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2641-1687
Research Article(ISSN: 2641-1687) 
Analysis of Association Between Rs3200401 Long NonCoding RNA MALAT1 Gene Polymorphism and Prostate Adenocarcinoma Development in Ukrainian Population Volume 1 - Issue 5
Volkogon Andrii1*, Chumachenko Yaroslav2, Harbuzova Viktoriia2 and Ataman Alexander3
- 1Department of Surgery and Oncology, Medical Institute of Sumy State University, Ukraine
- 2Scientific Laboratory of Molecular Genetic Research, Sumy State University, Ukraine
- 3Department of Physiology and Pathophysiology with the Course of Medical Biology, Medical Institute of Sumy State University, Ukraine
Received: May 10, 2019; Published: May 20, 2019
Corresponding author: Volkogon Andrii PhD, Department of Surgery and Oncology, Medical Institute of Sumy State University, Sumy, Ukraine
DOI: 10.32474/JUNS.2019.01.000122
Abstract
MALAT1 (metastatic associated lung adenocarcinoma transcript 1) is one of the most well-known and most conserved long non-coding RNA (lncRNA). This lncRNA is considered to be the biomarker of PC increased risk, and high expression of its gene correlates with the worst survival rate in patients with PC. Thus, the objective of current study is to analyze the possible link between rs3200401 locus of lncRNA MALAT1 gene and prostate adenocarcinoma (PA) development in Ukrainian population. Venous blood of 184 patients with PA and 66 men without cancer was used for case-control study. MALAT1 rs3200401 polymorphism genotyping was performed by real-time PCR method. The statistical analysis of obtained data was performed using SPSS 17.0. Our data showed that MALAT1 rs3200401 SNP was related to PA, regardless of adjustment for age, smoking status and body mass index. The risk of PA development was significantly lower in rs3200401T-alleles homozygote compared to C-allele carriers (OR = 0.164; P = 0.005) and C-allele homozygotes (OR = 0.170; Pa = 0.006). The presented work is the first study about the role of rs3200401 MALAT1 gene single nucleotide polymorphism in prostate cancer development. Obtained results suggested that MALAT1 rs3200401T-alleles can be the possible genetic risk factor for PA development in Ukrainian population.
Introduction
Today it is well known that protein-coding genes account for only 3% of the total active transcript. Careful analysis of other transcription products has shown the diversity of their structure and functions, as well as shed much light on the disclosure of their biological significance. To date, there are different approaches to classification of non-coding RNA (ncRNA), one of which is based on the length of their molecules [1,2]. Thus, non-coding RNAs that contain 200 or more nucleotides in their structure are called long non-coding RNA (lncRNA) and represent the largest fraction of all ncRNAs. In the living organism cells lncRNAs are involved in the processes of transcription, translation, epigenetic regulation of gene expression, inactivation of the X chromosome, cell differentiation, etc. [2-4]. One of the most well-known and most conserved lncRNAs is MALAT1 (metastatic associated lung adenocarcinoma transcript 1), also known as noncoding nuclear-enriched abundant transcript 2. MALAT1 gene is located on the plus-chain of the 11th chromosome (11q13.1), consists of more than 8,000 bases pairs and contains 2 exons [5]. The results of several studies have demonstrated the presence of MALAT1 in the nuclear paraspeckles, indicating its involvement in mRNA processing [6]. In addition, experiments with genetic knockout showed the importance of MALAT1 to attract the splicing factor SR, including SRF1 (Strubbelig-receptor family 1 protein) and SC35 (Serine/arginine-rich splicing factor SC35) to nuclear paraspeckles [7]. MALAT1 was firstly identified as metastasis prognostic factor in patients with non-small cell lung cancer [8], but more recent studies have demonstrated significant expression of this RNA in normal tissue cells (myocardium, renal epithelium, pulmonary epithelium, nerve tissue) [9]. It has been shown that MALAT1 enhanced expression leads to induction of cell proliferation by activating of ERK/MAPK and Wnt/β-catenin signaling pathways and may be involved in apoptosis of tumor cells, their migration and invasion [10]. A recent review by Amodio et al. [2]. showed the association between MALAT1 expression level and various types of malignant tumors such as lung cancer, stomach cancer, prostate cancer, ovarian cancer, bladder cancer, sarcoma and lymphoma [2]. The excessive MALAT1 expression in prostate cancer (PC) cells has also been demonstrated by MartensUzunova et al. [11]. In addition, this lncRNA is considered to be the biomarker of PC increased risk, and high expression of its gene correlates with the worst survival rate in patients with prostate and bladder cancer [12,13]. The link between MALAT1 genetic polymorphisms and different variants of malignant tumors is less studied. Wherein, studies of association between MALAT1 gene SNPs and PC risk are absent at all. Thus, the aim of current study is to analyze the possible link between rs3200401 locus of lncRNA MALAT1 gene and prostate adenocarcinoma (PA) development in Ukrainian population.
Materials and Methods
Venous blood of 184 patients with PA (mean age [±SD] 73.03±7.56 years) and 66 men (mean age 76.8±9.05 years) without cancer was used for case-control study. All patients were treated at the Sumy Regional Clinical Oncology Center. The final morphological diagnosis of PA was established in accordance with European Association of Urology Guidelines. All patients had II clinical stage of cancer according to TNM-classification of malignant tumors. The protocol was in accordance with the Helsinki Declaration and was approved by the Ethics Committee of the Medical Institute of Sumy State University (No. 4 / 05.18.09). Voluntary written informed consent was obtained from all participants. Venous blood sampling for genotyping was performed in sterile conditions in a 2.7ml Monovettes with 11.7mM EDTA addition (“Sarstedt”, Germany). DNA from blood leukocytes was extracted using GeneJET Whole Blood Genomic DNA Purification Mini Kit (ThermoFisher Scientific, USA). MALAT1 rs3200401 polymorphism genotyping was performed by real-time PCR method using 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, USA) and Taq-Man Assays (TaqMan®SNP Assay C_3246069_10). The amplification consisted of 50 cycles: initial denaturation – 95°C (20s), denaturation – 95 °C (30s) hybridization and elongation – 60.0 °C (30s). The data analysis was carried out using 7500 Fast Real-time PCR Software. The statistical analysis of obtained data was performed using SPSS (version 17.0, Chicago, IL, USA). To test the deviation of rs3200401 alleles from Hardy-Weinberg equilibrium and to compare the rs3200401 genotypes distribution between two groups χ2-Pearson criterion was used. Comparison of the mean values between two groups was performed using Student’s t-criterion (Shapiro-Wilk test was used to check the normality of distribution). To determine the risk of PA development odds ratio (OR) and 95% confidence interval (CI) were calculated for dominant, recessive, dominant, and additive models of inheritance. Such risk factors as age, smoking and body mass index (BMI) were used as covariates for multivariable logistic regression analysis. All tests were two-sided. P<0.05 was considered as significant.
Results
The clinical characteristics of 180 cases and 66 matched controls are presented in Table 1. It should be noted that the mean age of the control group was significantly higher than of the PA patients (P = 0.001). This circumstance allowed to increase the reliability of control, as the risk of oncological process development in representatives of this group in the further stages of their life was reduced. The comparison groups also differed in the number of smokers (P = 0.009) and did not differ in the mean value of BMI (P = 0.151). The distribution of MALAT1 rs3200401 alleles and genotypes in PA patients and control groups as well as the results of their comparison are shown in Table 2. It was revealed that alleles distribution in the control group was deviated from the Hardy-Weinberg equilibrium (P = 0.034). The comparative analysis of rs3200401 alleles distribution between PA patients and control individuals did not show a significant difference (P = 0.095). Instead, the difference in genotypes distribution between the comparison groups was statistically significant (P = 0.005). Table 3 presents the results of analysis of MALAT1 rs3200401 genotypic association with AP risk under different models of inheritance. Before adjustment for non-genetic risk factors the link between rs3200401 site and AP development was revealed for recessive and additive models. It was found that minor T-alleles homozygotes had significantly lower risk of prostate cancer development compared to C-allele carriers (ORc = 0.161; 95% CI = 0.047-0.555; Pc = 0.004) and C-allele homozygotes (ORc = 0.168; 95% CI = 0.048-0.589; Pc = 0.005). After adjusting for age, smoking status and BMI the overall picture of the results did not change: ORa = 0.164; 95% CI = 0.047-0.577; Pa = 0.005 – for recessive model; ORa = 0.170; 95% CI = 0.048-0.609; Pa = 0.006 – for additive model. The association between MALAT1 rs3200401 genotypes and risk of PA development under other models of inheritance was found neither before no after adjusting for covariates.
Table 1: Clinical characteristics of the comparison groups.
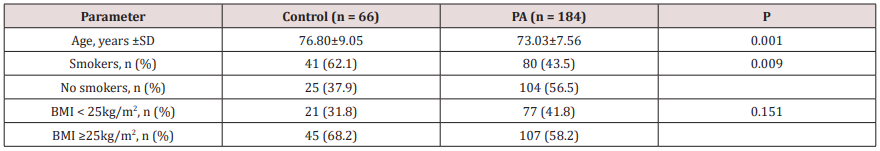
PA – prostate adenocarcinoma; n – number of people. The comparison of nominal variables was performed using the χ2 test, quantitative variables – using t-test.
Table 2: Clinical characteristics of the comparison groups.
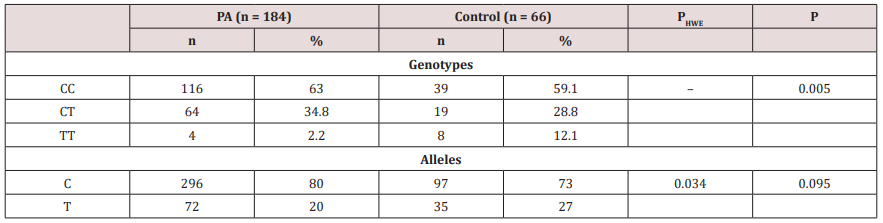
PA – prostate adenocarcinoma; n – number of people; PHWE – difference of alleles distribution from expected distribution by HardyWeinberg law; P – difference of alleles and genotypes distribution between comparison groups.
Table 3: Analysis of MALAT1 rs3200401 genotypic association with risk of PA development.

PA – prostate adenocarcinoma; 95% CI – confidence interval; Pc – crude P value (without adjusting for covariates); ORc – crude odds ratio; Pa – P value adjusted for age, smoking status and body mass index; ORa – adjusted odds ratio. A First row in additive model represents comparison of TT-genotype with CC-genotype, second row – comparison of CT-genotype with CC-genotype.
Discussion
The rs3200401 MALAT1 gene polymorphic site is the replacement of cytosine to thymine at 65504361-position of the plus chain of the 11th chromosome. There are three main variants of the MALAT1 transcripts, each of which has an appropriate placement of rs3200401 polymorphic locus: variant 1 (NR_002819.4) – 6624C>T, variant 2 (NR_144567.1) – 6390C>T and variant 3 (NR_144568.1) – 6147C>T. It was found that different regions of MALAT1 molecule have heterogeneous affinity for a number of molecules, which determines their involvement in a variety of regulatory processes. Miyagawa et al. [14] described two main fragments in the MALAT1 molecule – region E (nucleotides 1961- 3040) and region M (nucleotides 6008-7011) [14]. These regions determine the localization and functioning of MALAT1 in nuclear paraspeckles. Based on these data, Wang et al. [15] suggested that rs3200401 polymorphic site could affect the attachment of SC35 to the M-region of MALAT1. The results of experiments showed that rs3200401 C>T nucleotide replacement leads to increasing of MALAT1 molecule minimum free energy, which changes its structure and complicates its binding to SC35 factor. Violation of interaction between MALAT1 and SC35 ultimately leads to inhibition of pre-mRNA alternative splicing and expression of genes involved in tumors metastases. The authors concluded that such effect could explain both decreasing of tumor aggression activity and better survival rate in lung adenocarcinoma patients who are minor T-allele carriers [15]. The data obtained in our work indicate that there is a significant difference in MALAT1 rs3200401 genotypes distribution between PA patients and control group in Ukrainian population. It was revealed that subjects with TT-genotype have significantly lower risk of PA development compared to carriers of C-allele. To date, there is a small number of works devoted to the study of MALAT1 rs3200401 SNP role in various oncological processes development. At the same time, most of these studies results fully coincide with data obtained in our work. So, Peng et al. [16] showed that CT-heterozygotes have lower risk of breast cancer incidence compared to CC-homozygotes. Interestingly, haplotype analysis showed that Crs3200401Grs619586Grs7027113 variant carriers also have the low risk of breast cancer development [16]. Wang et al. [15] demonstrated that lung adenocarcinoma patients with rs3200401T-allele have significantly longer median survival compared to CC-genotype carriers. In addition, there was no association of this polymorphic site with survival of patients with skin squamous cell carcinoma [15]. It should be noted that our study has several limitations that should be taken into account for more relevant results evaluation. It was found that MALAT1 rs3200401 alleles distribution in the control group did not correspond to Hardy-Weinberg equilibrium. It could be explained by the fact that for this study we have selected only male individuals from general control group, which cannot fully reflect the real situation in the context of the entire population. It should also be noted that in order to make a firm conclusion about the link of rs3200401 locus with PA development more male individuals should be involved in both the control and case groups.
Conclusion
The presented work is the first study about the role of rs3200401 MALAT1 gene polymorphic site in prostate cancer development both in Ukrainian population and worldwide. The obtained results showed that rs3200401 locus was associated with the prostate adenocarcinoma onset in Ukrainian men. TTgenotype carriers have the lower risk of prostate adenocarcinoma development compared to C-alleles carriers.
References
- Jalali S, Kapoor S, Sivadas A (2015) Computational approaches towards understanding human long non-coding RNA biology. Bioinformatics 31(14): 2241-2251.
- Amodio N, Raimondi L, Juli G (2018) MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol 11(1): 63.
- Taft RJ, Pang KC, Mercer TR (2010) Non-coding RNAs: regulators of disease. J Pathol 220(2): 126-139.
- Akhade VS, Pal D, Kanduri C (2017) Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv Exp Med Biol 1008: 47-74.
- Gutschner T, Hämmerle M, Diederichs S (2013) MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 91(7): 791-801.
- Lin Y, Schmidt B, Bruchez M (2018) Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res 46(7): 3742-3752.
- Tripathi V, Ellis J, Shen Z (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39(6): 925-938.
- Ji P, Diederichs S, Wang W (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22(39): 8031-8041.
- Wu X, Wang X, Wu W (2014) MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther 15(6): 806-814.
- Danyang R, Huiying L, Renqiu Li (2016) Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications (Review). Oncol Lett 11(3): 1621-1630.
- Martens-Uzunova E, Böttcher R, Croce C (2014) Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 65(6): 1140-1151.
- Smolle M, Bauernhofer T, Pummer K (2017) Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer. Int J Mol Sci 18(2): 473.
- Li C, Cui Y, Liu L (2017) High Expression of Long Noncoding RNA MALAT1 Indicates a Poor Prognosis and Promotes Clinical Progression and Metastasis in Bladder Cancer. Clin Genitourin Cancer 15(5): 570-576.
- Miyagawa R, Tano K, Mizuno R (20120 Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18(4): 738-751.
- Wang J, Xiang J, Wu L (2017) A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer 17(1): 167.
- Peng R, Luo C, Guo Q (2018) Association analyses of genetic variants in long non-coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene 642: 241-248.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...