Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2643-6760
Research article(ISSN: 2643-6760) 
Quantification of MicroRNAs for the Diagnostic Screening of Colon Cancer in Human Stool by Absolute Digital(d)PCR* Volume 1 - Issue 5
Farid E Ahmed1*, Mostafa M Gouda2,3, and Nancy C Ahmed1
- 1 Institute for Research in Biotechnology, Greenville, USA
- 2Department of Nutrition, El-Bohooth Street, Dokki, Cairo, Egypt
- 3National Research and Development Center for Egg Processing, Wuhan, Hubei, PR, China
Received: January 08, 2019; Published: January 18, 2019
Corresponding author: Farid E Ahmed, Institute for Research in Biotechnology, 2905 South Memorial Drive, Greenville, NC 27834, USA
DOI: 10.32474/SCSOAJ.2019.01.000125
Abstract
There is currently no validated micro(mi)RNA diagnostic stool test to screen for colon cancer (CC) on the market because of the complexity of fecal density, vulnerability of stool to daily changes, and the presence of three sources of miRNAs in stool (cell-free from fecal homogenates, exsosomal miRNAs from fecal exosomes, and fecal colonocytes). By employing earlier on a microarray miRNA experiment, using Affymetrix GeneChip miRNA 2.0 Arrays, on immunocaptured and enriched stool colonocytes of 15 subjects [three healthy controls and twelve colon cancer patients [three TNM stage 0-1 (e.g., polyps 1 cm, villous or tubvillous, or with high grade dysplasia), three stage 2, three stage 3, and three stage 4] in triplicates, this allowed for selection of a smaller panel of 14 preferentially expressed mature miRNAs associated with colon cancer (12 Up-Regulated, miR-19a, miR-20a, miR-21, miR-31, miR-34a, miR-96, miR-106a, miR-133a, miR-135b, miR-206, miR-224 and miR-302; and 2 Down-Regulated, miR-143 and miR-145). Then carrying out an absolute quantitative digital PCR on these 15 stool samples from TNM stages 0-4 on total small RNA extracted by immunocapture, followed by RT that employed a Custom TaqMan® miRNA Reverse Transcription (RT) Kit and TaqMan RT Primer Pool, and absolute quantification of miRNAs, in copies/μl, measured using a chip-based Absolute QuantStudio 3D Digital PCR analysis, allowed for validating the microarray results. To ensure that human and not bacterial small total RNA was chosen, coextraction protocols with E. coli K1 strain RS18 was carried out, followed by comparing Agilent electrophoretic patterns with human and bacterial electrophoretic patterns, and also random samples were sequenced using mRNA/miRNA sequencing, to ensure that human and not bacterial mRNA was chosen.
Introduction
Quantitative dPCR miRNA data presented in herein, show that the quantitative changes in the expression of a few mature miRNA genes in stool, which are associated with right and left colon cancer, would provide for a more convenient, sensitive and specific diagnostic screening molecular markers, more useful than markers currently available on the market, such as the low-sensitivity (<15%) fecal occult blood test(FOBT); result in better compliance; and is more economical than the invasive and expensive colon cancer colonoscopy exam, resulting in a higher probability of curing that cancer, if detected at the early TNM stages, and which becomes incurable and deadly if not diagnosed before metastasis.
Advantages of Using a MiRNA Diagnostic Colon Cancer Screening Test
The expression of individual genes may be altered by mutations in the DNA, or by a change in their regulation at the RNA or protein levels [1]. Epigenetic silencing is an important mechanism that contributes to gene inactivation in colorectal cancer (CRC) [2]. Analysis of promoter methylation of hypermethylated in cancer 1 (HIC1) gene in human stool showed it to be highly specific (98%) for both colon adenoma and carcinoma [3], but the sensitivity was quite low (31% for adenoma & 42% for all cancer), suggesting that an epigenetic marker only is not adequate for an accurate diagnostic screening, but a combination of genetic and epigenetic markers would be required to reliably identify CRC at an early disease stage [4]. Working with the stable DNA has been relatively easy compared to working with the fragile RNA molecule [1].A study by scientists at Exact Sciences Corp, Marlborough, MA, which markets a mutation-based DNA test “Cologuard”, assessed a newer version of a fecal DNA test for CRC screening using a vimentin methylation marker and another mutation DY marker plus nondegraded DNA in a limited sample of 44 CRC patients and 122 normal controls [5].
It cited a sensitivity of 88% and a specificity of 82%, only for advanced cancer, but not for the early adenoma stage. Besides, DNA mutation tests are not cost-effective, as screening for multiple mutations is expensive because these demanding mutation tests are not automated and are labor intensive. In addition, mutation detection in oncogenes and suppressor genes suffers from: a) the detection of mutations in these genes in fewer than half of large adenomas and carcinomas, b) the detection of gene mutations in non-neoplastic tissues, c) mutations found only in a portion of the tumor, and d) mutations often produce changes in the expression of many other genes [6,7]. Protein-based methods are currently not suited for screening and early diagnosis, either because proteins are not specific to one tumor or tissue type (e.g., CEA), their susceptibility to proteases, current lack of means to amplify proteins, no function is known for more than 75% of predicted proteins of multicellular organisms, there is not always a direct correlation between protein abundance and activity, and most importantly because detection of these markers exfoliately often signifies the presence of an advanced tumor stage. The dynamic range of protein expression in minimally-invasive body fluids (e.g., blood) is as large as 1010. Moreover, mRNA levels do not necessarily correlate with protein expressions.
Protein microarray studies revealed that protein expression vastly exceeds RNA levels, and only post translationally modified proteins are involved in signal transduction pathways leading to tumorigenesis. There is no well-documented protein test that has been shown in clinical trials to be a sensitive and a specific indicator of colon neoplasia, especially in early stages [8]. A serum proteomic study employing liquid chromatography (LC)-mass spectrometry (MS) carried out in a non-biased fashion failed to differentiate between individuals with large adenoma ( 1 cm) and normal individuals [9]. Compared to nucleic acids, proteomic research is a newer discipline; therefore, it will take considerable time to identify and validate proteins suitable for use as clinical markers, and resolve issues of bias and validations [10].On the other hand, a transcriptomic mRNA approach, has been shown to detect both adenomas and colon carcinomas with high sensitivity and specificity in preliminary studies [1], but no randomized, standardized, blinded prospective clinical studies have been carried out to validate the superiority of the mRNA approach.
A study indicated that a combination of a transcriptomic mRNA and miRNA expression signatures improves biomolecular classification of CRC [11]. Furthermore, not only does miRNAs regulate mRNA, but they also regulate protein expression. Two studies have shown that a single miRNA act as a rheostat to fine tune the expression of hundreds of proteins [12,13]. Hence, for CRC screening, miRNA markers are much more comprehensive and preferable to a DNA-, epigenetic-, mRNA- or a protein-based marker [14-18]. An added advantage for the use of the stable, nondegradable miRNAs by PCR expression, by chip-based methods, is its being automatable, making them much more economical and more easily acceptable by laboratory personnel performing these assays [4]. The discovery of small non-coding protein sequences,17-27 nucleotides long RNAs (microRNAs), has opened new opportunities for developing a non-invasive screening test for early diagnosis of many cancers. The latest miRbase release 22 on, March 12, 2018 [http://ww.mirbase.org] indicates the total number of miRNAs labeled “high confidence” has increased by 168, to 1996, than in the previous release [19].
MiRNA functions seem regulate development [20], apoptosis [21], and specific miRNAs are essential in oncogenesis [22,23], effective in classifying solid [24-26] and liquid tumors [27,28], and could serve as oncogenes or suppressor genes [29]. MiRNA genes are frequently found at fragile sites, as well as minimal regions of loss of heterozygosity, or amplification of common break-point regions [30], implying their involvement in carcinogenesis. MiRNAs have potential to serve as biomarkers for cancer diagnosis, prognosis and/or response to therapy [31,32]. Profiles of miRNA expression differ between normal and tumor tissues (33,34), suggesting that their expression profiles cluster similar tumor types together more accurately than expression profiles of protein-coding mRNA genes [33,34]. A study that examined global expression of 735 miRNAs in 315 samples of normal colonic mucosa, tubulovillus adenomas, adenocarcinomas proficient in DNA mismatch repair (pMMR), and defective in DNA mismatch repair (dMMR) representing sporadic and inherited CRC stages I-IV suggest involvement of common biologic pathways in pMMR and dMMR tumors in spite of the presence of numerous molecular differences between them, including differences at the miRNA level; indicating the need to pay attention to mismatch DNA repair (MMR) [34] .
Unlike screening for large numbers of messenger (m)RNA [1], a modest number of miRNAs is used to differentiate cancer from normal [35], and unlike mRNA, miRNAs in stool remain largely intact and stable for detection [36], therefore, leading to conclude that miRNA molecules are better markers to use for developing a reliable noninvasive diagnostic marker screen for colon cancer [14-18], since: a) the presence of the bacterium Escherichia coli does not hinder detection of miRNA by a sensitive technique such as dPCR [36], and b) the miRNA expression patterns are the same in primary tumor, or diseased tissue, as in stool samples [35-37]. The gold standard to which the miRNA test is compared to, has been “colonoscopy”, obtained from patients’ medical records [38]. However, because the low sensitivity guaiac FOBT is still the most commonly used screen in annual checkups (www.cancer.org) [39], this test should also be included for comparison with the proposed dPCR diagnostic miRNA screening approach in human stool.
Advantages of Stool Over Other Testing Media
Stool testing has several advantages over other colon cancer screening methods a s it is truly noninvasive and requires no unpleasant cathartic preparation, formal health care visits, or time away from work or routine activities [40-43]. Unlike sigmoidoscopy, it reflects the full length of the colorectum and samples can be taken in a way that represents the right and left side of the colon. It is also believed that colonocytes are released continuously and abundantly into the fecal stream, contrary to blood that is released intermittently as in guaiac fecal occult blood test (FOBT) [39]; therefore, this natural enrichment phenomenon partially obviates the need to use a laboratory-enrichment technique to enrich for tumorigenic colonocytes, as for example when blood is used for screen testing [44]. Furthermore, because testing can be performed on mail-in-specimens, geographic access stool screening is essentially unimpeded [4]. The American Cancer Society (ACS) has recognized stool-based molecular testing as a promising screening technology for CRC (www.cancer.org).
Isolation of colonocytes from stool, and comparing the Agilent electrophoretic (18S and 28S) patterns to those obtained from total RNA extracted from whole stool [45-47], and differential lysis of colonocytes by RT lysis buffer (Qiagen), could be construed as a validation that the electrophoretic pattern observed in stool (18S and 28S) is truly due to the presence of human colonocytes, and not due to stool contamination with Escherichia coli (16S and 23S). Taking into account that some exsosomal RNA will be released from purified colonocytes into stool, attempts must be made to correct for exsosomal RNA effect [48].
MiRNA dPCR Study Design
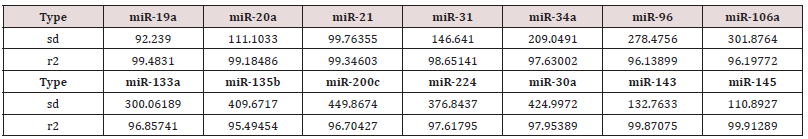
To test miRNAs as reliable, quantitative, sensitive and specific diagnostic biomarkers, for early non-invasive screening of colon cancer, using absolute dPCR test, preliminary work must be validated in a study using a nested case control epidemiology design and employing a prospective specimen collection, retrospective blind evaluation (Probe) of control subjects and test colon cancer patients, as specifically delineated by the National Cancer Institute’s Early Detection Research Network http://edrn.nci.nih.gov for cancer biomarker discovery studies. Selection of 14 miRNAs, 12 of them showed increased expression and 2 showed decreased expression for analysis of absolute miRNAs expression by a chipbased digital (d) PCR test is presented in Tables 1 & 2, and Figure 1. Absolute Quantitative Digital PCR Approach Digital PCR is a new approach to miRNAs quantification that offers an alternate method to qPCR for absolute quantification, by partitioning a sample of DNA or cDNA into many individual, parallel PCR reactions; some of these reactions contain the target molecule (positive), while others do not (negative).
Figure 1: Diagram illustrating QuantStudioTM 3D Digital PCR System Chip; ChipCase Lid (1); Digital PCR 20K 10 mm2 nanofluidic v2 chip (2), which contains 20,000 reaction wells; QuantStudioTM 3D Digital PCR Chip Case (3); Chip ID (4); Fill port (5); and Reaction wells, the 20,000 physical holes that suspend individual PCR reactions.

Table 2: Absolute Quantification of Up-/Down- Regulated miRNAs in Stool by QuantStudioTM 3D Chip-Based Digital PCR.
A single molecule can be amplified a million-fold or more. During amplification, TaqMan chemistry with dye-labeled probes is used to detect sequence-specific targets. When no target sequence is present, no signal accumulates. Following PCR analysis, the fraction of negative reactions is used to generate an absolute count of the number of target molecules in the sample, without the need for standards or endogenous controls. In conventional qPCR, the signal from wild-type sequences dominates and obscures the signal from rare sequences [49-51]. By minimizing the effect of competition between targets, dPCR overcomes the difficulties inherent to amplifying rare sequences and allows for sensitive & precise absolute quantification of the selected miRNAs. Applied Biosystem QuantStudio™ 3D instrument used in this research study, only performs the imaging and primary analysis of the digital chips. The chips themselves must be cycled offline on a Dual Flat Block GeneAmp® 9700 PCR System. or the ProFlex™ 2x Flat PCR System. The QuantStudio™ 3D Digital PCR System can read the digital chip in less than 1 minute, following thermal cycling [45].
It allows for one sample per chip; although, duplexing allows for analsis of two targets per chip. Sample prep for digital PCR is no different than for real-time qPCR, when using the QuantStudio™ 3D Digital PCR System. To figure out the concentration of cDNA stock from results, if one includes all of the necessary dilution factors into the AnalysisSuite™ software, the software will give the copies/μL in the stock. There are 2 dilutions that one needs to take into account:
a) The first is the dilution of the sample in the reaction,. and
b) The second is the dilution of the stock that one makes before adding it to the digital PCR reaction.
For example, if one wants to add 1 μL of a sample that has been diluted 1:10 from the stock. Thus, if one adds 1 μL of his/her sample to a 16 μL (final volume) reaction, the dilution factor of the sample is 1:16 or 1/16 = 0.0625. Since the stock has also been diluted 1:10 (0.1), one also need to factor this in. The final dilution factor to enter into the software is 0.0625 x 0.1 = 0.00625 (1:160). One can use either annotation to indicate the dilution factor in the AnalysisSuite™ software. If one enters that value into the “Dilution” column, the software will give the copies/μL in the starting material (stock).
The Poisson Plus algorithm for projects that contain QuantStudio™ 3D Chips with target, quantities >2000 copies/μL. The Poisson Plus algorithm corrects for well-to-well load volume variation, on a per Chip basis. This becomes important at higher target concentrations. There is also an option to export the Chip data as XML on the Export tab-thousands of discrete subunits prior to amplification by PCR, each ideally containing either zero or one (or at most, a few) template molecules [50]. Each partition behaves as an individual PCR reactions –as with real-time PCR-fluorescent FAM probes [or others, as VIC fluorescence]. Samples containing amplified products are considered positive (1, fluorescent), and those without product –with little or no fluorescence (i.e., are negative, 0). The ratio of positives to negatives in each sample is the basis of amplification. Unlike real-time qPCR, dPCR does not rely on the number of amplification cycles to determine the initial amount of template nucleic acid in each sample, but it relies on Poisson Statistics to determine the absolute template quantity.
The unique sample partitioning step of dPCR, coupled with Poisson Statistics allows for higher precision than both traditional end point PCR. and qPCR methods; thereby allowing for analysis of rare miRNA targets quantitatively and accuratley [47,48]. The use of a nanofluidic chip, shown below, provides a convenient and straight forward mechanism to run thousands of PCR reactions in parallel. Each well is loaded with a mixture of sample, master mix, and Applied Biosystems TaqMan Assay reagents, and individually analyzed to detect the presence (positive) or absence (negative) of an endpoint signal. To account for wells that may have received more than one molecule of the target sequence, a correction factor is applied using the Poisson model. It features a filter set that is optimized for the FAM™, VIC®, and ROX™ dyes, available from Life Technologies [46]. The chips themselves must be cycled offline on a Dual Flat Block GeneAmp® 9700 PCR System. or the ProFlex™ 2x Flat PCR System. The QuantStudio™ 3D Digital PCR System can read the digital chip in less than 1 minute, following thermal cycling [45].
It allows for one sample per chip; although, duplexing allows for analsis of two targets per chip. Sample prep for digital PCR is no different than for real-time PCR, when using the Quant Studio™ 3D Digital PCR System. To figure out the concentration of cDNA stock from results, if one includes all of the necessary dilution factors into the AnalysisSuite™ software, the software will give the copies/sL in the stock. There are 2 dilutions that one needs to take into account:
a) The first is the dilution of the sample in the reaction,. and
b) The second is the dilution of the stock that one makes before adding it to the digital PCR reaction.
For example, if one wants to add 1 μL of a sample that has been diluted 1:10 from the stock. Thus, if one adds 1 μL of his/her sample to a 16 μL (final volume) reaction, the dilution factor of the sample is 1:16 or 1/16 = 0.0625. Since the stock has also been diluted 1:10 (0.1), one also need to factor this in. The final dilution factor to enter into the software is 0.0625 x 0.1 = 0.00625 (1:160). One can use either annotation to indicate the dilution factor in the AnalysisSuite™ software.
If one enters that value into the “Dilution” column, the software will give the copies/μL in the starting material (stock). The Poisson Plus algorithm for projects that contain Quant Studio™ 3D Chips with target, quantities >2000 copies/μL. The Poisson Plus algorithm corrects for well-to-well load volume variation, on a per Chip basis. This becomes important at higher target concentrations. There is also an option to export the Chip data as XML on the Export tab-thousands of discrete subunits prior to amplification by PCR, each ideally containing either zero or one (or at most, a few) template molecules [47]. Each partition behaves as an individual PCR reactions –as with real-time PCR—fluorescent FAM probes [or others, as VIC fluorescence]. Samples containing amplified products are considered positive (1, fluorescent), and those without product –with little or no fluorescence (i.e., are negative, 0). The ratio of positives to negatives in each sample is the basis of amplification. Unlike real-time qPCR, dPCR does not rely on the number of amplification cycles to determine the initial amount of template nucleic acid in each sample, but it relies on Poisson Statistics to determine the absolute template quantity.
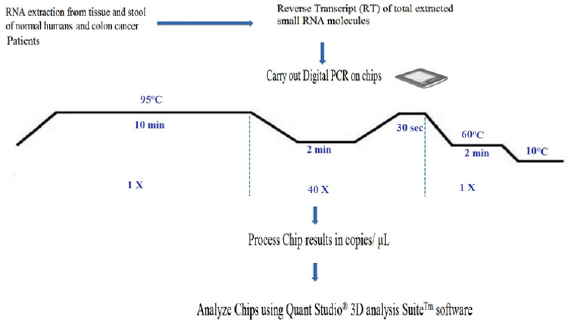
The unique sample partitioning step of dPCR, coupled with Poisson Statistics allows for higher precision than both traditional and qPCR methods; thereby allowing for analysis of rare miRNA targets quantitatively and accuratley [47]. The use of a nanofluidic chip, shown below, provides a convenient and straight forward mechanism to run thousands of PCR reactions in parallel. Each well is loaded with a mixture of sample, master mix, and Applied Biosystems TaqMan Assay reagents, and individually analyzed to detect the presence (positive) or absence (negative) of an endpoint signal. To account for wells that may have received more than one molecule of the target sequence, a correction factor is applied using the Poisson model. It features a filter set that is optimized for the FAM™, VIC®, and ROX™ dyes, available from Life Technologies [46]. A workflow of the dPCR procedure by the QuantStudioTM 3D Digital PCR System is presented in (Figure 2) Workflow of a digital miRNAs PCR for colon cancer profiling in human colon tissue or stool samples.
Figure 2: Workflow of a digital miRNAs PCR for colon cancer profiling in human colon tissue or stool samples.
Digital PCR, however, has Several Tips to Follow:
i. A rough estimate of the concentration of miRNAs of interest has to be first carried out, in order to make appropriate dilutions, so that not too many partitions will get multiple copies that prevent accurate calculation of the copy number of miRNAs of interest;
ii. Non-template controls and a RT negative control must be set up for each miRNA, when using a “primer pool method” for retro-transcription;
iii. A chip-based dPCR method requires less pipetting steps, which reduces potential PCR contamination compared to another type of dPCR marketed by Bio-Rad Laboratories, thus called “Bio-Rad’s droplet digital PCR”, which requires multiple pipette transfers that potentially increase the risk of contamination [47], and
iv. Quant StudioTM 3D chip has 20,000 fixed reaction wells, whereas Bio-Rad’s droplet PCR relies upon the generation of droplets;
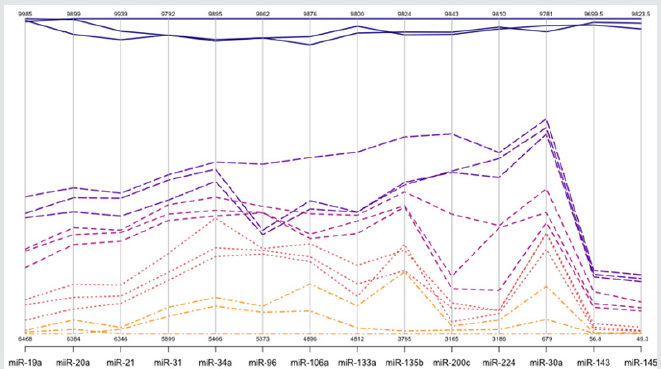
a step that could be extremely variable, as reported by Miotto et al (11,48) Absolute dPCR data tabulated in Tables 1 & 2, and presented graphically in Figure 1 below, which show 14 preferentially expressed mature miRNAs associated with colon cancer (12 Up-Regulated, miR-19a, miR-20a, miR-21, miR-31, miR- 34a, miR-96, miR-106a, miR-133a, miR-135b, miR-206, miR-224 and miR-302; and 2 Down-Regulated, miR-143 and miR-145) in stool samples from healthy controls, and stages 0-1 to 4 individuals with colon cancer . Standard deviations (sd) obtained from the one-way ANOVA, using the 5 level factor Type (normal, stage01, stage2, stage3, stage4) were calculated. The adjusted R-squared values representing the proportion of variation explained by Type are also reported. Type was statistically significant for every gene; all p-values were less than 0.000001 (no adjustments for multiple comparisons). These data are tabulated in Table 3 and shown graphically in Figure 1. For each gene on the graph in Figure 1, the min and max have been shown, in order to make the presentation clearer. At top left is high exxpression Value of 9985, which is the maximum value for that gene, at the bottom one finds the value for the minimum the colors range from dark blue (control) to orange (stage 4).
The groups are also distinguished by line type: control (solid), stage 0-1 (long dash), stage 2 (dash), stage 3 (dot), stage 4 (dash nd dot). The figure is a parallel coordinate plot made in R, using the package MASS. For statistical analysis. Innovation and Clininical Significance of the dPCR-miRNA Diagnostic Stool Screening Approach Innovation lies in the collective use of many methods, such as: immunoparamagnetic beads to capture colonocytes from the harsh, noninvasive stool environment, whose extracted fragile total small RNA is stabilized shortly after stool excretion by commercial kits so it does not ever fragment, followed by standardized analytical quantitative miRNA dPCR-chip profiling in stool samples, which are neither labor intensive, nor require extensive sample preparation, to develop a panel of few stable miRNAs for absolute quantitative diagnostic screening of early sporadic colon cancer (stage 0-1), cheaper, with higher sensitivity and specificity than any other colon cancer screening test on the market . Isolation of colonocytes from stool samples is needed to provide a quantitative estimate of how our proposed miRNA method performs. Although we may miss exosome RNA, a parallel test could also be carried out on miRNAs obtained from stool samples to compare the extent of loss when colonocytes are only used, and an appropriate correction for exsosomal loss can be made [48]. Figure 3 Absolute Quantification of Up- or Down-Regulated miRNAs in Human Stool by Quant Studio TM 3D Digital PCR Chip System.
Figure 3: Absolute Quantification of Up- or Down-Regulated miRNAs in Human Stool by QuantStudioTM 3D Digital PCR Chip System.
Acknowledgments
We express our thanks to participating volunteers who provided stool samples for the study; Dr. Paul W. Vos of the Department of Biostatistics, East Carolina University for Statistical Analysis, and Dr. Clark D. Jeffries at Renaissance Computing Institute, University of North Carolina at Chapel Hill for his insight on bioinformatics’ analysis.
References
- Ahmed FE, Vos P, iJames S, Lysle DT, Allison RR, et al. (2007) Transcriptomic molecular markers for screening human colon cancer in stool & tissue. Cancer Genom Proteom 4(1) : 1-20.
- Ahmed FE (2007) Colon cancer epigenetics: The role of environmental factors and the search for molecular biomarkers. J Env Sci Health C 25(2): 101-154.
- Lenhard K, Bommer GT, Asutay S, Schauer R, Brabletz T, et al. (2005) Analysis of promoter methylation in stool: a novel method for the detection of colorectal cancer. Clin Gastroenterol Hepatol 3(2): 142-149.
- Davies RJ, Miller R and Coleman N (2005) Colorectal cancer screening: prospects for molecular stool analysis. Nature Rev Cancer 5(3): 199-209.
- Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC, et al. (2007) Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol 5(1): 111-117.
- Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA and Ross MA (2004) Fecal DNA versus fecal occult blood for colorectal cancer screening in an average-risk population. New Eng J Med 351(26): 2704- 2714.
- Cairns R and Sidaransky D (1999) Molecular methods for diagnosis of cancer. Biochim Biophys Acta 1423(2): 11-18.
- Ahmed FE (2009) Liquid chromatography-mass spectrometry: A tool for proteome analysis & biomarker discovery and validation. Exp Opin Mol Diag 3(4): 429-444.
- Greenbaum D, Colangelo C, Williams K and Gerstein M (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 4(9): 117-126.
- Rifai N, Gillette MA, Carr SA (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechol 24(8): 971-983.
- Lanza G, Ferracin M, Gafa R, Veronese A, Spizzo R, et al. (2007) mRNA/ microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 6: 54.
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, et al. (2008) The impact of microRNA on protein output. Nature 455(7209): 64-71.
- Selbach M, Schwarheuser B, Thierfelder N, Fang Z, Khanin R, et al. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58-63.
- Lanza G, Ferracin M, Gafa R, Veronese A, Spizzo R, et al. (2007) mRNA/ microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 6: 54.
- Ahmed FE, Ahmed NC, Gouda M and Vos PW (2017) MiRNAs for the Diagnostic Screening of Early Stages of Colon Cancer in Stool or Blood. Surgical Case Reports and Reviewss 1(1): 1-19
- Ahmed FE, (2014) miRNA as markers for the diagnostic screening of colon cancer. Expert Rev Anticancer Ther.14(4): 463-485.
- Ahmed FE, Ahmed NC, Gouda MC. and Bonnerup C (2017) MicroRNA as molecular markers for screening of colon cancer. Case Reports Surg Invasive Procedures 1(2): 14-15.
- Ahmed FE (2017) MicroRNAs as Molecular Markers for Colon Cancer Diagnostic Screening in Stool & Blood. Int Med Rev 9: 124.
- Kozomara A and Griffiths Jones S (2014) miRBase: annotating high confidence microRNA using deep-sequencing data. Nucleic Acids Res 42: 68-73.
- Reinhart BJ, Slack FJ, Basson M, Pasquinell AE, Bettinger JC, et.al. (2000) RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772): 901-906.
- Xu P, Guo M and Hay BA (2004) MicroRNAs and the regulation of cell death. Trend Genet 20(12): 617-624.
- Gregory RI and Shiekhattar R (2005) MicroRNA biogenesis and cancer. Cancer Res 65(9): 3509-3512.
- Cummins JM and Velculescu VE (2006) Implication of microRNA profiling for cancer diagnosis. Oncogene 25(46): 6220-6227.
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, et al. (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9(3): 189-198.
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, et al. (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65(16): 7065-7070.
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA Jr, et al. (2006) The colorectal microRNome. Proc Natl Acad Sci USA 103(10): 3687-3692.
- Calin GA, Ferracin M, Cimmino A, Dileva G, Shimiz M, et al. (2005) A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Eng J Med 353(17): 1793-1801.
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, et al. (2003) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102(10): 3627-3632.
- Chang-Zheng C (2005) MicroRNAs as oncogenes and tumor supressors. N Eng J Med 353(17): 1768-1771.
- Calin GA, Sevignai C, Dumitru CD, Hyslop T, Noch E, et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101(9): 2999-3004.
- Schepler T, Reinert JT, Oslenfeld MS, Christensen LL, Silahtaroglu AN, et al. (2008) Diagnostic and prognostic microRNAs in Stage II colon cancer. Cancer Res 68(15): 6416-6424.
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, et al. (2008) MicroRNA expression profile associated with progression and therapeutic outcome in colon adenocarcinoma. J Am Med Assoc 299(4): 425-436.
- Barbarotto E, Schmittgen TD and Calin GA (2008) MicroRNAs and cancer: Profile, profile, profile. Int J Cancer 122(5): 969-977.
- Calin GA and Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857-866.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435(7403): 834-838.
- Oberg AL, French AJ, French AJ, Sarver AL, Subramanian S, et al. (2011) MiRNA expression in colon polyps provide evidence for a multihit model of colon cancer. PLoS ONE 6(6).
- Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, et al. (2009) Diagnostic microRNA markers for screening sporadic human colon cancer and ulcerative colitis in stool and tissue. Cancer Genom Proteom 6(5): 281- 296.
- Ahmed FE (2007) The role of microRNA in carcinogenesis and biomarker selection: a methodological perspective. Exp Rev Mol Diag 7(5): 569- 603.
- Ahmed FE (2014) Role of microRNA molecules in colon cancer etiology. Biol Med 6: 201
- Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, et al. (2013) Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics10(3): 93-113.
- Peterson NB, Murff HJ, Ness RM and Dittus RS (2007) Colorectal cancer screening among men and women in the United States. J. Womens Health 16(1): 57-65.
- Morikawa T, Kato J, Yamaji, Wada R, Miksushima T, et al. (2006) Comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology 129: 422-428.
- Matsushita HM, Matsumura Y, Moriya Y, Akasu T, Fujita S, et al. (2005) A new method for isolating colonocytes from naturally evacuated feces and its clinical application to colorectal cancer diagnosis. Gastroenterology 126: 1918-1927.
- Davidson LA, Lupton JR, Miskovsky E, Fields AP and Chapkin RS (2003) Quantification of human intestinal gene expression profiling using exfoliated colonocytes: a pilot study. Biomarkers 8(1): 51-61.
- Jorgensen OD, Kronborg O and Fenger C (2002) A randomized study of screening for colorectal cancer using faecal occult blood testing: results after 13 years and seven biennial screening rounds. Gut 50(1): 29-32.
- Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, et al. (2010) Fecal microRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev 19(7):1766-1774.
- Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, et al. (2012) Diagnostic microRNA markers to screen for sporadic human colon cancer in blood. Cancer Genom Proteom 9(4): 179-192.
- RNA Integrity Number (RIN)-Standardization of RNA Quality Control Agilent Application Note Publication Number 5989-1165EN016.
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, et al. (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology 7: 3.
- Fleige S, and Pfaffl MW (2006) RNA integrity and the effect on the realtime qRT-PCR performance. Molecular Aspects of Medicine 27(2-3): 126-139.
- Valadi H, Elkstrom K, Bossios A, Sjostrand M, Lee JJ, et al. (2007) Exosome mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654-659.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...