Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2644-1373
Research ArticleOpen Access
The Anti-Inflammatory Activity of Ferulic Acid on NF- κBDepends on Keap1 Volume 2 - Issue 2
Miriam Giacomarra, Annalisa Mangano and Giovanna Montana*
- Istituto di Ricerca e Innovazione Biomedica, Consiglio Nazionale delle Ricerche, Via Ugo La Malfa 153, 90146 Palermo, Italy
Received: August 14, 2020; Published: August 27, 2020
Corresponding author: Giovanna Montana, Istituto di Ricerca e InnovazioneBiomedica, Consiglio Nazionale delleRicerche, Via Ugo La Malfa 153, 90146 Palermo, Italy
DOI: 10.32474/LOJPCR.2020.02.000133
Abstract
Nrf2 and NF-κB are the two master transcriptional factors activated by different cellular signals turned to counteract the deleterious effects of pathological cellular processes linked to inflammation and oxidative stress.Several recent studies have highlighted a molecular connection between NF-κB and Keap1/Nrf2 pathways. The Keap1 protein seems to be the central player in this interaction, as it is involved in both IKKβ-NF-κB and Nrf2 modulation. Ferulic acid (FA) is a well-known antioxidant and antiinflammatory agent, able to relieveinflammatory response viaNF-κB/IKK kinase, but until now the complete molecular network under its action is not all clear. Immunoblot data conducted on LPS-treated macrophage-like RAW264.7 cells transfected with si- Keap1 show that the FA anti-inflammatory and modulatory effects on NF-κB are abolished. Luciferase assay conducted in human A549 cell line, in which Keap1 protein is partially inactive, highlights that NF-κB activation induced by LPS is refractory to FA inhibition. This study proved that Keap1 and IKK together are important modulators of NF-κB and their activity is essential for FA anti-inflammatory effectiveness.
Introduction
The process of inflammation leads to the onset of a state of oxidative stress and a series of cascade reactions that are associated with chronic diseases such as cancer, autoimmune disorders, and metabolic diseases [1]. NF-κB is the master transcriptional factor mainly involved in the activation of inflammation and immunity, whose excessive upregulation is associated with many human diseases, including inflammatory disease and cancer [2,3]. External pro-inflammatory stimuli activate NF-κB that induces the expression and the release of a set of inflammatory mediators such as IL-6, a pleiotropic cytokine contributing to switch the acute to chronic phase of inflammation [4,5]. NF-κB is mainly regulated by IKK kinase, which in presence of pro-inflammatory stimuli phosphorylates the inhibitory protein IkB, that dissociates from NF-κB with subsequent nuclear translocation [6]. Recently, many studies have highlighted the interaction between NF-κB/ IKK kinase and Keap1protein [7,8]. Our previous studies showed that Keap1 is a modulator both of Nrf2 and NF-κB pathways [9]. In physiological conditions, Keap1 maintains Nrf2 levels low. In conditions of oxidative stress, Keap1 is oxidized on the reactive cysteine residues and inactivated, so that Nrf2 moves into the nucleus. Nrf2 (nuclear factor erythroid2-related factor2), belongs to the basic leucine zipper (bZIP) transcription factor and heterodimerizes with small Maf proteins [10], and it is the primary player in the inducible cell defense system.
It binds to the promoter region of genes involved in redox regulation, proteostasis, DNA repair, prevention of apoptosis, iron and heme metabolism, and phase I, II, and III drug/xenobiotic metabolism [11]. The activation of this factor is controlled at the transcriptional and post-transcriptional level through the regulation of its stability, the post-transcriptional changes, and the availability of its binding partners [12,13]. In response to different stimuli, Nrf2 moves to the nucleus where it activates the transcription of its target genes, such as HO-1. Nrf2 is a modular protein presenting seven domains of homology to Nrf2-ECH (Neh), each of which performs a different function. In particular, the Neh2 domain binds Keap1 in the Kelch repeats [14]. In addition, Keap1 has five major domains: an N-terminal broad complex, tram track, and bric-a-brac (BTB) domain; a central intervening region (IVR) and a series of six C-terminal Kelch repeats.
The IVR and BTB domains are required for the redox-sensitive regulation of Nrf2 through a series of reactive cysteines. The 624 amino acids of murine Keap1 include 25 cysteines [15,16]. All cysteine residues are conserved and cysteines C257, C273, C288, and C297, located in the intervening region (IVR) domain, give Keap1 the molecular sensor able to respond to such a diverse array of chemical stimuli [17]. Early studies have identified IKKβ and Keap1 as the key drivers of inflammatory response [18,19]. IKK kinase is the main target of several anti-inflammatory molecules and kinase inhibitors are very effective in the control of many diseases [20-23]. In fact, LPS-signalling that is activated in macrophages via TLR4 involves a number of kinases. After binding to its receptor, the signal is transmitted by different adapter proteins: TIRAP (Tollinterleukin 1 receptor domain-containing adapter protein) and MyD88 (Myeloid differentiation primary response gene 88) driving the MyD88-dependent pathway, while TRAM (TRIF-related adaptor molecule) and TRIF (TIR-domain-containing adapter-inducing interferon-β) drive the MyD88-independent pathway.
The MyD88-dependent pathway converges into IKK (IκB kinase) and MAPK (Mitogen-activated protein kinases) activation and subsequent activation of NF-κB and activator protein AP- 1. In LPS-activated THP-1 cells, a human monocytic cell line, has been reported thanks to bioactive compounds such as flavonoids, hydroxycinnamic acids, tannins and in particular ferulic acid (FA), a hydroxycinnamic acid derivative, an effective inhibition of NF-κB activation as well as a decrease in the expression of proinflammatory cytokines TNF-α and IL-1β [24,25]. Furthermore, FA has a wide range of therapeutic effects like anti-inflammatory, antidiabetic, neuroprotective and hepatoprotective properties in others cellular models and in animals [26]. The present study carried out in RAW264.7 cells shows that the ferulic acid exerts its anti-inflammatory activity when Keap1 and IKK kinase are functionally active.
Materials and Methods
The mouse macrophage-like virus-transformed leukemia cell line RAW 264.7 was purchased from American Type Culture Collection (ATCC). RAW 264.7 cells and A549 cell line (provided by Francesca Sardina e Cinzia Rinaldo (IBPM-CNR, Roma, Italy) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% v/v heat-inactivated fetal bovine serum (FBS) and antibiotics (100U/ml penicillin and 100μg/ml streptomycin) at 37 °C in a humidified atmosphere with 5% CO2. FBS, DMEM, penicillin and streptomycin (10,000 U/ml) were purchased from GIBCO (Grand Island, NY). LPS from E. coli serotype O55: B5 and ferulic acid (FA; CAS Number: 537-98-4) were purchased from Sigma-Aldrich, Inc. (St. Louis, MO). TRIzol was purchased from Invitrogen. The Quanti Nova RT PCR kit and Quanti Nova Sybr Green PCR kit were obtained from Qiagen (Hilden, Germany). Nitrocellulose Blotting Membrane was purchased from Amersham Protran (Buckinghamshire, UK).
RT-Qpcr
RAW 264.7 cells were cultured (1 ×106 cells/well) in a 6-well plate overnight. Cells were treated with 100 ng/ml LPS or without (negative control) in the presence or absence of 100μM ferulic acid in DMEM supplemented with 10% bovine serum for 4 hours. Cells stimulated with 100ng/ml LPS for 4 hours served as a positive control. After such stimulation, the cells were detached from the wells and washed once with PBS. Total RNA was isolated with TRIzol according to the manufacturer’s instructions and was quantified by UV absorbance spectrophotometry and reverse transcribed with Quanti Nova RT PCR kit. QPCR was performed in triplicate on each cDNA sample for each gene:
IKK-β NM 010546 F:5’-GCCAGGGAGACTTGATGG-3’ R:5’-
GAGGTCTGTGCTTTAGCTGCTT-3’
IL6 NM 031168 F:5’-GAGGATACCACTCCCAACAGACC -3’ R:5’-
AAGTGCATCATCGTTGTTCATACA - 3’
HO-1 NM 0125082 F:5’ -GCGAAACAAGCAGAACCCA-3’ R:5’ –
GCTCAGGATGAGTACCTCCCA- 3’
HPRT NM 194F:5′-GCTATAAATTCTTTGCTGACCTGCTG-3′ R:
5′-AATTACTTTTATGTCCCCTGTTGACTGG-3′
by using primers set Quantitect from Qiagen. The threshold cycle (CT) values were calculated against the housekeeping gene HPRT. At least three distinct biological samples were examined for each gene and treatment (each one performed in triplicate). The cycling parameters were set as 95 °C for 5min, followed by 40 cycles at 95 °C 15sec and 60 °C for 2-min. The expression was calculated by the 2-ΔΔCt.
Transfection and luciferase reporter assays
A549 cells (3 × 104 cells/well) were seeded in 96-well plates and allowed to adhere for 24 hours. The cells were then cotransfected with pKEAP1Vector, pIL-6FL and Renilla-Luc plasmids (Promega) using the Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After that, the cell culture medium was removed and replaced with fresh medium containing 100μM of FA for 1-hour, followed by co-incubation with 100ng/ml of LPS for 4 hours. Then, the cells were harvested, the luciferase activity was quantified by using the Dual-Glo Luciferase Assay System (Promega) and was normalised according to Renilla luciferase activity. The level of induction was calculated as the ratio of the normalised luciferase activity of LPS stimulated cells compared to non-stimulated cells and of association between LPS and FA compared to treatment with agents alone. Relative light units were measured in a GloMax 96 microplate luminometer (Promega).
Sirna transfection
The RAW264.7 cells (5×105 cells/well) were seeded in 6-well plates for 24 hours. Briefly, the siRNA pool for Keap1 (Qiagen) and NC-siRNA (Qiagen) were incubated with Lipofectamine RNAiMAX (Promega) in basal media with no serum or antibiotics and allowed to complex for 10 min at room temperature. Then, the complex was added to the cell suspension of each well (final siRNA pool concentration of 10nM). Finally, cells were incubated for 24 hours in a humidified incubator and then used for the analysis.
Western blotting
RAW 264.7 cells (1x107 cells) were cultured in 10-cm dishes (Falcon) and allowed to adhere for 24 hours. After treatment with FA 100μM 1 followed by co-incubation with LPS 100ngr/ml for 4 hours, the cells were washed twice with cold PBS and lysed in lysis buffer (10mM Hepes pH 7.9, 10mM KCl, 1.5mM MgCl2, 0.1mM EGTA pH 7, 0.05 mM DDT, and 1% protease inhibitor cocktail (SIGMA). After vortexing for 10s, the lysates were centrifuged at 1250g for 15min; the supernatants (cytoplasmic extract) were immediately transferred to a clean pre-chilled tube and put on ice. The pelleted nuclei were re-suspended in a hypertonic buffer (5% glycerol, 1.5mM MgCl2, 0.1mM EGTA, 0.05mM DTT, 0.4 M NaCl, 20mM PMSF and 10mM HEPES pH 7.9) and shaken for 60min at 4 °C. The proteins in the supernatants (nuclear extract) were collected by centrifugation at 20817g for 15min, then immediately transferred to a clean pre-chilled tube and put on ice. Whole cell lysates were obtained using RIPA buffer (Cell Signalling Inc. Beverly, MA, USA). The protein concentration of cell lysates was determined by the Bradford method.
An equivalent amount of protein (30μg) from whole or nuclear and cytoplasm fractions, respectively, was separated on 8-16% Tris-Glycine Gel (BioRad) gels by electrophoresis and transferred to a nitrocellulose membrane. The membranes were subsequently incubated for 1 h at room temperature with 3% BSA in TBS buffer (0.1% v/v) to block non-specific binding and incubated with an appropriate primary antibody in 1% BSA in TBST (tween 0.01% v/v). Antibodies polyclonal anti-mouse recognizing p-IKKα/β, IKKα/β, p65 NF-κB, lamin B1, Keap1, Nrf2, Histone H3 and betaactin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Incubation with the secondary antibodies Alexa Fluor 680 goat anti-rabbit (1:2000) and Alexa Fluor 800 rabbit anti-mouse (1:5000) (Molecular Probes, Life Technologies, Carlsbad, CA, USA) was performed for 1h at room temperature. Densitometry analysis was conducted using the Odyssey Infrared Imaging System (Li-COR Bioscience, Nebraska, USA).
Statistics
Statistical analysis and graphical presentation were performed using the statistical package GraphPad Prism Version 8 (GraphPad Software, Inc,USA). Data were expressed as mean ± SD and were evaluated using unpaired t-test calculator or one-way analysis of variance (ANOVA) for multiple comparisons. The differences were considered statistically significant when p values were <0.05.
Results
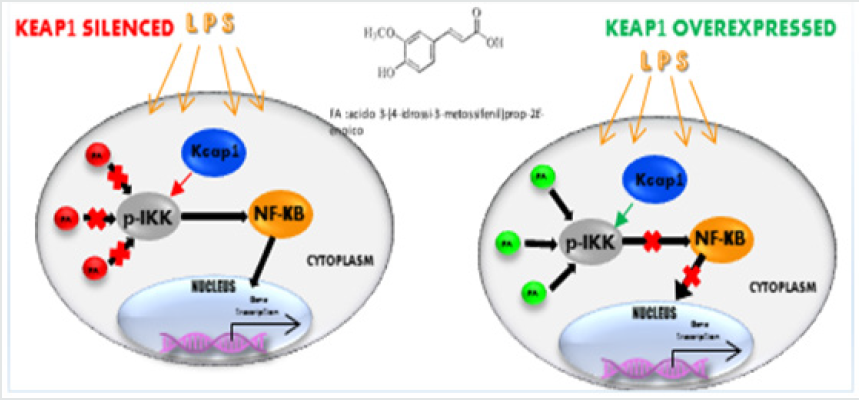
Keap1 silenced abolishes ferulic acid modulation on NF- κB-IKK pathway in LPS-treated RAW 264.7 cells
Our previous study [9] has proved that in Keap1- silenced RAW264.7 cells, the LPS treatment induced a higher proinflammatory cytokines mRNA expression compared with ncsiRNA transfected cells and moreover FA treatment resulted not able to reduce cytokines mRNA expression induced by LPS. Other authors [27] showed that depletion of Nrf2 induced the activation of NF-κB and a higher expression of TNF-α, IL-1β, IL-6 cytokines. In addition, it was reported that Keap1 is identified as an IKKβ interacting protein, involved in IKKβ phosphorylation [28]. To verify a potential Keap1 role in the FA modulation on NF-κB pathway, we analyzed by Western blot analysis NF-κB-IKK pathway in depleted Keap1 and FA-LPS treated RAW 264.7 cells. Cells were transfected with siRNAs for Keap1 and, after 24h, exposed to 100ng/ml LPS for 4h. One hour before LPS, cells were treated or not with 100μM FA, a known inhibitor of the NF-κB-signalling pathway in LPS-activated RAW 264.7 [29]. Cytoplasmic and nuclear extracts were prepared and analyzed in immunoblotting for the NF-κB p65. Figure 1A (panel a) shows that in Keap1-silenced and LPS-treated cells there is a significant increase of p65 in the nuclear fraction, compared to non LPS treated cells, and in accordance with our previous evidences [9], pre-treatment with FA does not decrease NF-κB translocation. In Figure 1A (panel b) is showed Western blot analysis of NF-κB pathway in nc siRNA transfected and FA-LPS treated RAW 264.7 cells, where FA is able to modulate p65 translocation also when LPS treatment occurs.
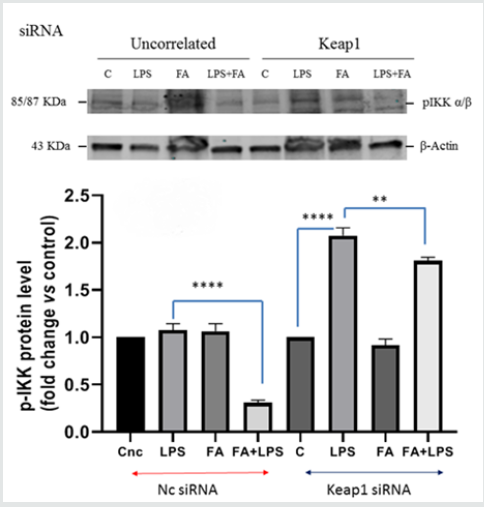
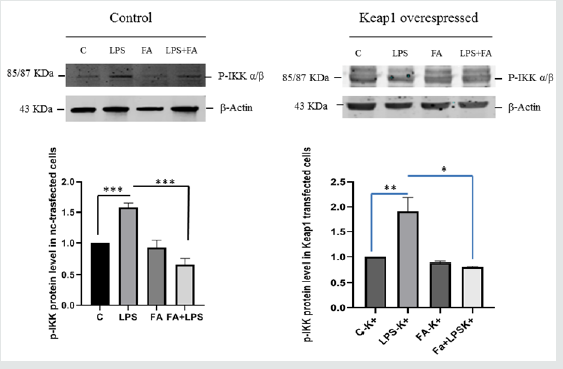
IKK kinase activates NF-kB pathway, so we analyzed whether Keap1 silencing had any influence on the kinase expression and activation. Total extract protein was prepared and analyzed in immunoblotting for IKKα/β. As reported in Figure 1B, when LPS treatment occurs, in the nc-siRNA transfected RAW264.7 cells, there is an increase of IKK level protein which decreases when cells were pre-treated with FA (p<0.0005). Instead, in the Keap1- silenced RAW 264.7 cells, neither LPS treatment nor FA have effect. QPCR analysis of IKKβ mRNA expression was conducted on total RNA extracted from Keap1 silenced and LPS treated RAW 264.7 cells. The results showed that IKKβ mRNA expression is lower (1,9-fold LPS vs C, p<0.0001) than the nc-transfected cells, where there is an increase of 8,4 fold LPS vs c (Figure 1C). These data indicate that Keap1-silencing has effect on IKKβ transcription and protein expression. Moreover, we investigated the Keap1 silencing influence on IKKα/β phosphorylation. The results are showed in Figure 1D: in nc-silenced and LPS treated cells, FA inhibits IKK phosphorylation (0,3fold FA+LPS vs 1,08 fold LPS), not such as in Keap1 silenced cells (1,8-fold FA+LPS vs 2 fold LPS). The results show that the Keap1 knockdown results in a much-altered IKK pathway modulation by FA.
Figure 1A:Keap1 silencing and ferulic acid modulation on NF-κB pathway in LPS-stimulated RAW 264.7 cells. RAW264.7 cells transfected with si-Keap1were treated with or without FA 100μM for 1 h, then treated with LPS 100 ngr/ml for 4h like the uncorrelated siRNA transfected RAW264.7 cells.(Panel a).Immunoblot analysis of p65 translocation in RAW264.7 cells transfected with si-Keap1. The relative protein levels of NF-κB p65 in the nucleus and in the cytoplasm are shown in the graph. Difference between means FA+LPS vs LPS in RAW264.7 cells transfected with si-Keap1±SEM 0,03667±0,1119 is not significantly different (P<0.05), instead difference between means FA+LPS vs C±SEM 0,7833±0,09144 is significantly different (P<0.05), such as the difference between means LPS vs C±SEM0,7467±0,02011.
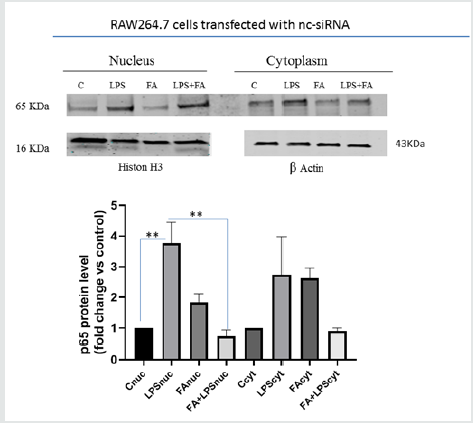
Figure 1A:Keap1 silencing and ferulic acid modulation on NF-κB pathway in LPS-stimulated RAW 264.7 cells. RAW264.7 cells transfected with si-Keap1were treated with or without FA 100μM for 1h, then treated with LPS 100ngr/ml for 4h like the uncorrelated siRNA transfected RAW264.7 cells.(Panel b).Immunoblot analysis of p65 translocation in RAW264.7 cells transfected with nc- si-RNA. Cells were treated as described above and the relative p65 protein level in the nucleus and in the cytoplasm is shown in the graph. Difference between the means (LPS-C) ± SEM 2,767 ± 0,3930 is significantly different(p<0.05). Difference between the means (FA+LPS -LPS) ± SEM-3,033 ± 0,4110 is also significantly different (p<0.05). Cytoplasmic and nuclear extracts were immunoblotted with an anti-β-actin or anti-Lamin B and H3 antibody, respectively.The data shown represent three independent experiments.
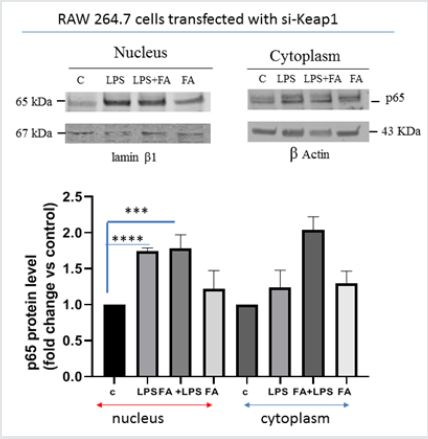
Figure 1B: Analysis of IKK kinase protein level in RAW264.7 cells Keap1 silenced. RAW264.7 cells were treated as described before. After treatment, cells were harvested and lysed. Total proteins were extracted and were analyzed by immunoblot. Difference between means (FA+LPSsiKeap1-LPSsiKeap1)±SEM is not significantly different (P<0.05). In RAW264.7 transfected with nc-siRNA the difference between means (FA+LPS-LPS)±SEM -50,10±1,238 (p<0.05) is significantly different. β-actin immunolabelling was used as loading control.The data shown represents three independent experiments.
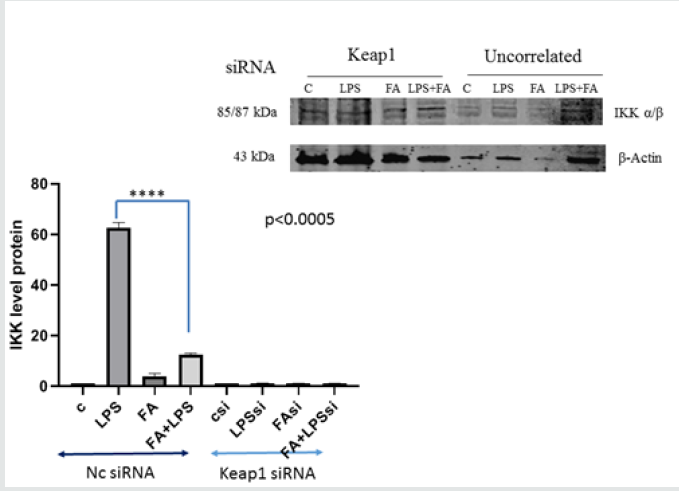
Figure 1C: IKKβ mRNA expression in LPS nc-siRNA and in LPS siKeap1-transfected RAW 264.7 cells. IKKβ mRNA relative quantity was analyzed by qPCR in cells treated with LPS 100ng/ml for 4 hours; total RNA was extracted, and cDNA was analyzed by qPCR. Non-stimulated cells worked as a negative control. The graph shows the level of mRNA expression in LPS nc-siRNA transfected cells and in LPS siKeap1-transfected cells. Difference between means (LPSnc-LPSsiKeap1) ± SEM-6,483 ± 0,2709 is significantly different (P < 0.05).The data shown represents three independent experiments each of which performed in triplicate.
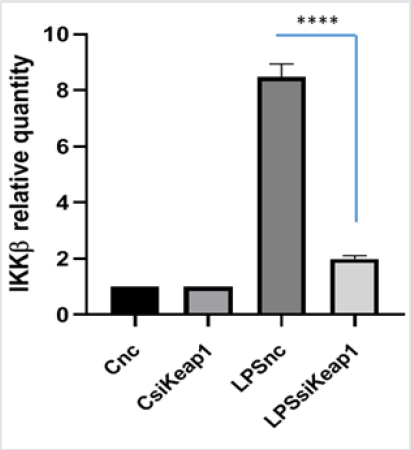
Figure 1D: IKKα/β phosphorylation induced by LPS in Keap1silenced cells. RAW264.7 cells were treated as described before. After treatment, cells were harvested and lysed; total proteins were extracted, and equal amounts of proteins were analyzed for phospho- IKKα/β by immunoblotting analysis. The expression level of p-IKKα/β is shown in the graph. Difference between means (LPSsiKeap1-CsiKeap1)±SEM 1,073 ± 0,04807 is significantly different (p<0.05).Difference between means (FA+LPSsiKeap1-LPSsiKeap1)±SEM-0,2633±0,05239 issignificantly different (p<0.05). Difference between means (LPSnc- Cnc) ± SEM 0,07667 ± 0,03930 is not significantly different(p<0.05). Difference between means (FA+LPSnc-LPSnc)±SEM -0,7700±0,04308 is significantly different (p<0.05). β-actin immunolabelling was used as a loading control.The data shown represents three independent experiments.
Ferulic acid is effectiveness on NF-κB activation in RAW 264.7cells Keap1 overexpressed
The overexpression of a specific protein is a common method for investigating the specific biological function and the mechanism of action. In order to confirm the data observed previously, we decided to over-express Keap1and to analyze the FA modulation on NF-κB/IKKβ activation. In our previous paper we described the FA effectiveness on inhibition of NF-κB activity [29]. We transfected RAW 264.7 cells with the expression vector for Keap1, let the cells grow for 24 hours, conducted the treatments as already described and extracted total RNA. IL6 is the major inflammatory gene under the NF-κB control, so to examine the effect of Keap1 overexpression on NF-κB modulation by FA, we analyze IL6 mRNA expression in LPS- treated and Keap1- overexpressed cells. Interestingly, FA reduces IL6 mRNA expression such as in RAW 264.7 transfected with pGEM-vector as control and LPS treated cells (Figure 2A). These results show that Keap1 overexpression does not alter neither the NF-κB activation neither the anti-inflammatory effect of FA. To confirm this result, we verify the p65 translocation occurring in LPS treated and Keap1 overexpressed cells in Figure 2A. We conducted an immunoblot analysis on proteins extracted from nuclear and cytoplasmic fractions. Figure 2B shows that LPS induces p65 translocation and FA-modulation occurs (0,5fold FA+LPS vs 6,8-fold LPS p<0.0001), unlike in Keap1 silencing cells. We have also analyzed IKKα/β phosphorylation. As showed in Figure 2C, in Keap1 overexpressed and LPS treated cells, FA is able to modulate IKK phosphorylation. The results show that the Keap1 overexpression does not alter the ferulic acid anti-inflammatory effect on NF-κb pathway.
Figure 2A:Analysis of Il6 mRNA expression in RAW 264.7 cellstransfected with the expression vector for Keap1 and pGEMvector as control. RAW 264.7 cells were transfected and after 24h cells were pre-treated with or without FA 100 μM for 1h and then treated with LPS 100ngr/ml for 4h; total RNA was extracted, and cDNA was analyzed by qPCR. The graph shows the level of IL6 mRNA expression in cells transfected with vector control and in cells transfected with Keap1-vector. Difference between means (FA+LPSk+-LPSk+)±SEM -1,917±0,4877 is significantly different (P<0.05) such asLPSk+-Ck+ (1,690±0,4004). Difference between means (FA+LPSnc-LPSnc)±SEM-15,09 ± 1,571 is significantly different (P<0.05). The data shown represents three independent experiments each of which performed in triplicate.
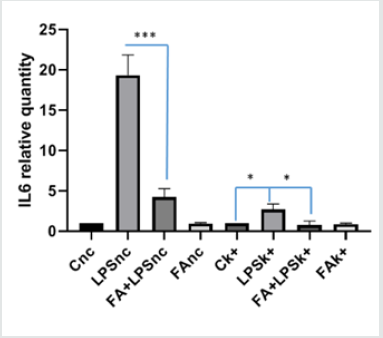
Figure 2B: Cytoplasmic and nuclear protein were extracted and immunoblot analysis of p65 was carried out. The graph shows the relative changes in protein level. Difference between means (FA+LPSnuc - LPSnuc)±SEM-6,327±0,09315 is significantly different (P<0.05). Difference between means (LPSnuc - Cnuc)±SEM 5,827±0,08090 is significantly different (P<0.05).Histon H3 and β-Actin immunolabelling were used as loading controls for nuclear and cytoplasmic fractions, respectively.The data shown represents three independent experiments.
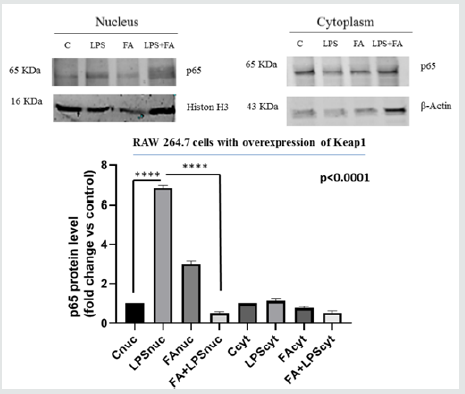
Figure 2C: Analysis of IKK phosphorylation in Keap1 overexpressed RAW264.7 cells. The cells were treated as described above. Total proteins were extracted from RAW 264.7 cells transfected with the expression vector for Keap1 and from RAW 264.7 cells transfected with pGEM-vector as control. p-IKKα/β proteins were measured by western blotting. Relative changes in protein intensity were quantified for Keap1 overexpressed RAW264.7 cells (right), and for basal level Keap1 cells (left) by densitometric analysis. Difference between means (FA+LPSk+-LPSk+) ± SEM-1,095±0,2102 is significantly different (p<0.05). Difference between means (LPSk+-Ck+)±SEM 0,9100 ± 0,1626 issignificantly different (P<0.05).Difference between means (FA+LPSc-LPSc) ± SEM -0,9100 ± 0,06298 is significantly different (P<0.05). Difference between means (LPSc-Cc) ± SEM 0,5800 ± 0,04163 is significantly different (P<0.05). β-actin immunolabelling was used as a loading control. The data shown represents three independent experiments.
A549 cells a useful model to analyze FA effect on NF-kB activities
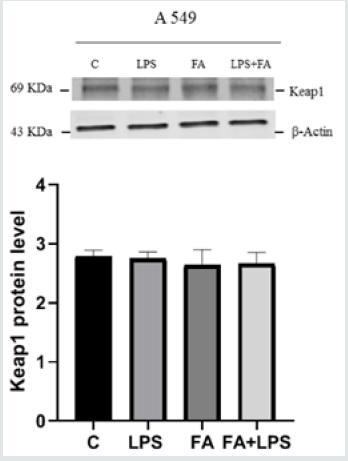
To confirm the results obtained by molecular approach by silencing the messenger for Keap1, we used A549 cell line. A549 is a lung carcinoma cell line that has an inactivating mutation on Keap1 allele, which translates into a protein with a minor binding capacity towards Nrf2 [30]. This cell line is a useful model to study Keap1 loss of functionality. We transfected A549 cells with pIL6- Luc, containing the IL6 promoter region and examined the effects of FA pre-treatment on luciferase activity and ferulic acid capability to alleviate inflammatory response via NF-κB pathway. Figure 3A shows that the up-regulation of luciferase activity LPS induced is not modulated by pre-treatment with FA (171fold FA+LPS vs 111fold LPS p<0.0001). Immunoblot for IKKα/β in Figure 3B shows that the effectiveness of FA is lost, in fact there is not a significant decrease of IKK protein level when LPS and FA treatment occurs. Moreover, to verify the level of Keap1 protein in A549 cells we conducted an Immunoblot on total proteins extracted from A549 treated or no with FA and LPS. Figure 3C shows that there are not significantly differences in Keap1 level in cells treated or not. These results follow the Keap1 silencing data indicate that Keap1and IKK are involved in the ferulic acid anti-inflammatory effectiveness.
Figure 3A:Analysis of ferulic acid modulation of IL6- promoter activity in A549 cells treated with LPS and FA. A549 cells (3×105 cells/well) were transfected with IL6-Luc reporter or vector for 24h and co-transfected with pRK-Renilla for internal normalization. Twenty-four hours after transfection, cells were pretreated with or without FA (100 μM) for 1h, then LPS was added (100ngr/ml). After 4h of activation, total cells were lysed and Luciferase activity was measured with a dual-luciferase system. The graph shows the relative light emission level of IL-6 promoter activation, expressed as difference between means±SEMof three experiments each one performed in triplicate.Difference between means (LPS+FA-LPS)±SEM 60,00±3,367, difference between means (LPS-C)±SEM 11,00±1,732 are significantly different (p<0.05).
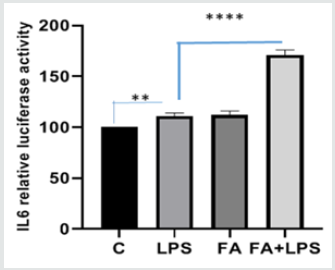
Figure 3B: IKK kinase protein level in A549 cell line. A549 cells were pre-treated with or without FA 100μM for 1h and then with LPS 100ngr/ml for 4h. Total proteins were extracted and immunoblot analysis of IKKα/β protein level was effectuated. Densitometric analysis is shown in the graph. Difference between means (FA+LPS-LPS) ± SEM 0,4167±0,1271 is significantly different (P < 0.05).The data shown represents three independent experiments. β-actin immunolabelling was used as a loading control
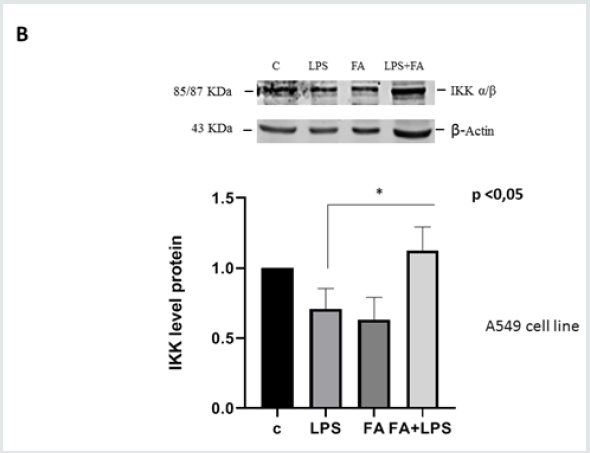
Figure 3C: Keap1 protein level in A549 cell line. A549 cells were treated as described above and total proteins were extracted and immunoblot analysis of Keap1 protein was conducted. The data shown represents three independent experiments.Analysis by one-way Anovashows thatSDs are not significantly different(P<0.05).
Ferulic acid maintains its anti-oxidant effectiveness on the Nrf2/HO-1 pathway modulation in Keap1silenced RAW264.7 cells
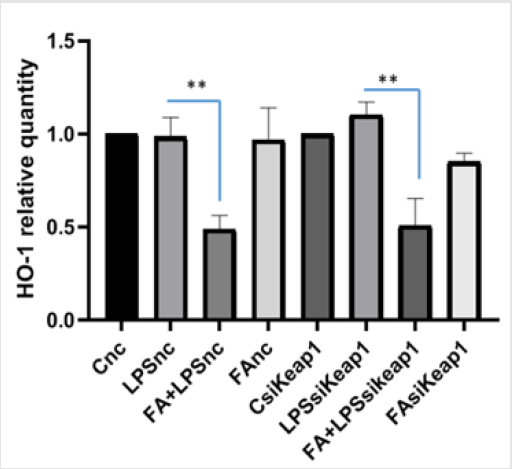
Nuclear transcription factor erythroid 2-related factor (Nrf2) is the most important transcription factor regulating the coordinated expression of antioxidant enzyme response such as Heme Oxygenase-1 (HO-1). Nrf2 resides in the cytoplasm by forming a complex with Keap1 that regulates its degradation by the proteasome. Our previous study carried out on LPS- treated RAW264.7 cells has shown that pre-treatment with FA determined a down-regulation of HO-1 protein [9]. Thus, to evaluate if FA maintains the antioxidant properties, also in altered Keap1 expression conditions, we analyzed the HO-1 expression in Keap1silenced RAW264.7 cells. As shown in Figure 4, FA down regulated the expression of HO-1 mRNA. These results indicate that FA maintains its antioxidant effect and the capability to modulate Nrf2 activity.
Figure 4: Keap1 silencing and ferulic acid antioxidant effect on Nrf2/HO-1 pathway. HO-1 mRNA expression was analyzed by qPCR in si-Keap1 RAW264.7 and nc-RNA transfected respectively. The cells were pretreated with or without FA 100μM for 1h and then with LPS 100ngr/ml for 4h. The graph shows the HO-1 relative quantity in Keap1 silenced RAW264.7 cells and nc-RNA transfected cells treated with FA and LPS. Difference between means (LPS+FAsiKeap1-LPSsiKeap1)±SEM -0,5963±0,09315 is significantly different (P < 0.05). In nc-RNA transfected cells the difference between means (FA+LPS-LPS) ± SEM-0,4967 ± 0,07520 is significantly different (P < 0.05).
Discussion
FA, a phenolic phytochemical found in many traditional Chinese medicines such as Angelica Radix and Chuanxiong Rhizoma, appears to be a potential therapeutic agent for treating various inflammatory disorders, although the molecular mechanism underlying the effectiveness is not entirely known. The pharmacological activity of FA in vascular endothelial cells is demonstrated, in fact, capable of inhibiting the expression of adhesion molecules in HUVECs [31]. Therefore, it was demonstrated that free and SLNs-loaded FA recover cell viability in neuroblastoma cells LAN5 [32]. FA treatment, in particular if loaded into SLNs, decreased ROS generation (Figure 5), restored mitochondrial membrane potential [Deltapsi(m)] and reduced cytochrome c release and intrinsic pathway apoptosis activation. In addition, FA modulated the expression of Peroxiredoxin, an anti-oxidative protein, and attenuated phosphorylation of ERK1/2 activated by Abeta oligomers. In our study highlight the Keap1 partnership in the molecular mechanism underlying the well-known anti-inflammatory effect of ferulic acid. Macrophage-like RAW264.7 stimulated by LPS was used in this study in that macrophages play vital roles in regulating inflammatory responses and LPS, the main component of the outer membrane of gram-negative bacteria, binding to the TLR4 receptor promotes inflammation [33]. Many data indicate that the molecular mechanisms underlying inflammation and oxidative stress are interconnected. NF-κB and Nrf2 are the main transcription factors on which a lot of antioxidant and inflammatory genes depend, but also tumorigenesis and apoptosis. There seems to be many the ways in which the two pathways interact, and Keap1 is one of the main [34]. Our results showed that Keap1 protein is the key point of the regulation carried out by FA on NF-κB pathway through IKK kinase. Recently, an ETGE motif (NQE36TGE39) similar to that of Nrf2 (DEE79TGE82) has been identified in IKKβ and so it is reasonable to suppose that Keap1 is an IKK modulator, regulating the ubiquitination and subsequent degradation [12,13,14].
Moreover, our data highlight that Keap1is able to influence the transcription and protein expression of IKKβ, but the mechanism is not clear. In fact, in Keap1 silenced RAW264.7 cells it is observed a decreased IKKβ transcription and protein expression in RAW264.7 when LPS treatment occurs. Remarkably, Keap1 silencing does not inactivate NF-κB pathway, but the FA modulation is lost. In our previous study, we demonstrated that IKK is an upstream target of ferulic acid in LPS-induced NF-κB activation [29]. It can be hypothesized that Keap1 plays an important role in the NF-κB/ IKK modulation FA mediated. Yet, to confirm the data observed previously, we overexpress Keap1 in RAW264.7 cells and we analyze FA modulation on NF-κB/IKK pathway after treatment with LPS. Immunoblot data confirm that LPS stimulus activates NF-κB/ IKK pathway and FA modulation occurs.
Therefore, Keap1 and IKK kinase proteins are very important for the FA effectiveness. To confirm the results obtained by molecular approach by silencing the messenger for Keap1, we used A549 cell line to examine the biological effect of Keap1 protein mutation on the NF-κB/IKK modulation by FA. A549, in fact, is a lung carcinoma cell line that has an inactivating mutation on Keap1 allele, and the level of the Keap1 protein is the same in non-treated and LPS treated cells. In this cell line the Nrf2 pathway is dysfunctional, because the NRF2-Keap1interaction is altered [30]. The Luciferase assay data show that LPS induces up-regulation of IL-6 promoter in LPS and in FA+LPS treated cells. It is probable so the Keap1 evolvement in NF-κB/IKK FA-modulation. Recent results have revealed that the cysteine residues Cys-151, Cys-273 and Cys-288 have been identified as direct sensors of electrophiles and oxidants [17]. Keap1 acts as a molecular sensor of ROS, NO and oxidant species and the interaction with oxidizing species results in an activation signal towards Nrf2. Nrf2 is an important mammalian member of the CncC family, which has been studied in numerous cellular systems and represents an important target for drug discovery in different diseases. Nrf2 is distributed in the cytoplasm together with Keap1 normally [12,13,14]. Under oxidative stress, Nrf2 was released and migrated to the nucleus, inducing HO-1 expression. HO-1, an antioxidant enzyme in the stress response, catalyzing the degradation of heme, biliverdin, and carbon monoxide (CO), also plays a key role in inflammation and oxidation responses.
Our previous study showed that the pre-treatment with FA carried out on LPS-treated RAW264.7 cells determined a decrease of HO-1 protein level [9]. Until now there are not any data demonstrating IKK involvement in Nrf2 modulation, so to verify the FA effectiveness and IKK/Keap1 involvement we analyze the HO-1 expression in Keap1silenced RAW264.7 cells. The results showed that pretreatment with FA decreased HO-1 mRNA expression in Keap1silenced RAW264.7 cells. Therefore, the results obtained agree with Kim et al [28] who identified Keap1 as IKK interacting protein and highlight that the Keap1 protein underlie the FA effectiveness on NF-κB modulation. The network Keap1-Nrf2-NF-κB/IKKβ is very interesting: it has many implications, being involved in inflammation, chemoresistance and cellular homeostasis [16,17,18]. Further studies are necessary to clarify the role of Keap1 in the NF-κB/ IKKβ dialogue. Our results also show that the Keap1 is an attractive target for therapeutic purposes, especially to improve the efficacy of anticancer and antiinflammatory molecules.
Conflict of Interest
The authors declare that there are no conflicts of interest.
Acknowledgements
Please Replace to Special thanks go to Diana Boraschi and Giovanni Viegi for their helpful suggestions and precious advice and Giovanni Duro to contributed to publication of this paper . I warmly thank Prof Francesco Mineo for the careful language revision.
References
- Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420(6917):860-867.
- Bonizzi G, Karin M (2004) The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 25(6):280-288
- Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr (2000) Selective activation of NF-kappa B subunits in human breast cancer: Potential roles for NF-kappa B2/p52 and for Bcl3. Oncogene 19(9):1123-1131.
- W Xiao, DR Hodge, L Wang, X Yang, X Zhang, et al. (2004) NF-kappaB activates IL-6 expression through cooperation with c-Jun and IL6-AP1 site, but is independent of its IL6-NFkappaB regulatory site in autocrine human multiple myeloma cells. Cancer Biol Ther 3(10):1007-1017.
- JV Castell, MJ Gomez Lechon, M David, T Hirano, T Kishimoto, et al. (1988) Recombinant human interleukin-6 (IL-6/BSF-2/HSF) regulates the synthesis of acute phase proteins in human hepatocytes. FEBS Lett 232(2):347-350.
- F Yang, E Tang, K Guan, CY Wang (2003) IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. The Journal of Immunology 170(11):5630-5645.
- Kanamoto M, Tsuchiya Y, Nakao Y, Suzuki T, Motohashi H, et al. (2018) Structural instability of IκB kinase β promotes autophagic degradation through enhancement of Keap 1 binding. PLoS One 13(11):e0203978.
- Lv P, Xue P, Dong J, Peng H, Clewell R, et al. (2013) Keap1 silencing boosts lipopolysaccharide-induced transcription of interleukin 6 via activation of nuclear factor κB in macrophages. Toxicol Appl Pharmacol 272(3):697-702.
- Lampiasi N, Montana G (2018) An in vitro inflammation model to study the Nrf2 and NF-κB crosstalk in presence of ferulic acid as modulator. Immunobiology 223(4-5):349-355.
- Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M (1995) Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol 15(8):4184-4193.
- Dinkova Kostova Albena T, Baird Liam, Holmström Kira M, Meyer Colin J, Abramov Andrey Y (2015) The spatiotemporal regulation of the Keap1-Nrf2 pathway and its importance in cellular bioenergetics. Biochem Soc Trans 43(4):602-610.
- Keum YS, Choi BY (2014) Molecular and chemical regulation of the Keap1-Nrf2 signaling pathway. Molecules 19(7):10074-10089.
- Nguyen T, Yang CS, Pickett CB (2004) The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radic Biol Med 37(4):433-441.
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD (2004) “Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem 279(30):31556-31567.
- Yamamoto T, Suzuki T, A Kobayashi, J Wakabayashi, J Maher, et al. (2008) Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Molecular and Cellular Biology 28:2758-2770.
- Karin Linnewiel Hermonia, Yair Motrob, Yifat Millerb, Joseph Levya, Yoav Sharoni (2014) Carotenoid derivatives inhibit nuclear factor kappa B activity in bone and cancer cells by targeting key thiol groups. Free Radical Biology and Medicine 75:105-120.
- Saito R, Suzuki T, Hiramoto K, Asami S, Naganuma E, et al. (2015) Characterizations of Three Major Cysteine Sensors of Keap1 in Stress Response. Mol Cell Biol 36(2):271-284.
- Syed Minhaj Uddin Ahmeda, Lin Luob, Akhileshwar Namania Xiu, Jun Wang, Xiuwen Tang (2017) Nrf2 signaling pathway: Pivotal roles in inflammation. Biochimi Biophys Acta Mol Basis Dis 1863(2):585-597.
- Wenjun Tu, Hong Wang, Song Li, Qiang Liu, Hong Sha (2019) The Anti-Inflammatory and Antioxidant Mechanisms of the Keap1/Nrf2/ARE Signaling Pathway in Chronic Diseases. Aging Dis 10(3):637-651.
- Long W, Liu P, Li Q, Xu Y, Gao J (2008) 3D-QSAR studies on a class of IKK-2 inhibitors with GALAHAD used to develop molecular alignment models. QSAR & Combinatorial Science 27(9):1113-1119.
- Christopher JA, Bamborough P, Alder C, Campbell A, Cutler GJ, et al. (2009) Discovery of 6-Aryl-7-alkoxyisoquinoline inhibitors of IB Kinase-(IKK-). Journal of Medical Chemistry 52(9):3098-3102.
- Nagarajan S, Doddareddy M, Choo H, Cho Y, Oh K, et al. (2009) IKK inhibitors identification. Part I: homology model assisted structure based virtual screening. Bioorg Med Chem 17:2759-2766.
- Avila CM, Romeiro NC, Sant’Anna CM, Barreiro EJ, Fraga CA (2009) Structural insights into IKK beta inhibition by natural products staurosporine and quercetin. Bioorganic & Medicinal Chemistry Letters 19(24):6907-6910.
- (2015) Molecules. 20:15319-15329.
- Srinivasan M, Sudheer AR, Menon VP (2007) Ferulic Acid: therapeutic potential through its antioxidant property. J Clin Biochem Nutr 40(2):92-100.
- Hao Pan, Handong Wang, Xiaoliang Wang, Lin Zhu, Lei Mao (2012) The absence of Nrf2 Enhances NF-κB-Dependent Inflammation following Scratch Injury in Mouse Primary Cultured Astrocytes. Mediators of Inflammation.
- Kim JE, You DJ, Lee C, Ahn C, Seong JY, et al. (2010) Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal 22(11):1645-1654.
- Lampiasi N, Montana G (2016) The molecular events behind ferulic acid mediated modulation of IL-6 expression in LPS-activated Raw 264.7 cells. Immunobiology 221(3):486-93.
- Anju Singh, Vikas Misra, Rajesh K, Thimmulappa, Hannah Lee (2009) Dysfunctional KEAP1-NRF2 Interaction in Non-Small-Cell Lung Cancer PLoS Med 3(10):e420.
- Zeng Chun MA, Qian HONG, Yu Guang WANG, Hong Ling TAN, Cheng Rong XIAO, et al. (2010) Ferulic Acid Attenuates Adhesion Molecule Expression in Gamma Radiated Human Umbilical Vascular Endothelial Cells. Biol Pharm Bull 33(5):752-758.
- Picone P, Bondi ML, Montana G, Bruno A, Pitarresi G, et al. (2009) Ferulic acid inhibits oxidative stress and cell death induced by Ab oligomers: improved delivery by solid lipid nanoparticles. Free Radic Res 43(11):1133-1145.
- Lu YC, Yeh WC, Ohashi PS (2008) LPS/TLR4 signal transduction pathway. Cytokine 42(2):145-151.
- Farzane Sivandzade, Shikha Prasad, Aditya Bhalerao, Luca Cucullo (2019) NRF2 and NF-қB interplay in cerebrovascular and neurodegenerative disorders: Molecular mechanisms and possible therapeutic approaches. Redox Biol 21:101059.
- Jeffrey R, Liddell (2017) Interplay between Nrf2 and NF-κB in Neuroinflammatory Diseases. Journal of Clinical Cellular Immunology 8:1-10.