Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2644-1373
Research ArticleOpen Access
Alignment of SARS-Cov-2 Spike Protein Compared with Ebola, and Herpes Simplex Viruses in Human Murine and Bats Volume 2 - Issue 3
Hend M Tag1,2*, Salwa S Bawazir1 and Ibtihal H Badawi1
- 1Department of Biology, College of Science and Arts at Khulais, University of Jeddah, Saudi Arabia
- 2Department of Zoology, Faculty of Sciences, Suez Canal University, Egypt
Received: February 08, 2021 Published: February 18, 2021
Corresponding author: Hend MT, Department of Biology and zoology, College of Science and Arts at Khulais, University of Jeddah, Jeddah, Saudi Arabia
DOI: 10.32474/LOJPCR.2021.02.000140
Abstract
Our analytical approach consisted in sequence alignment analysis of spike protein in different viruses, followed by construction of phylogenetic tree. Additionally, we investigated some commotional parameters on the protein sequence determining chemical composition as well as estimated PI .Our observation revealed significant difference in S protein between the 3 tested viruses in different species These differences may have significant implications on pathogenesis and entry to host cell.
Introduction
Viruses are inert outside the host cell which are unable to generate energy. As obligate intracellular parasites, during replication, they are fully reliant on the complex biochemical machinery of eukaryotic or prokaryotic cells. The central purpose of a virus is to transport its genome into the host cell to allow its expression by the host cell [1]. Binding of a single surface glycoprotein of virus to its host receptor promotes pH-dependent conformational variations once within the endosome acidic environment, thereby transporting the viral bilayer in closeness with the host cell membrane to promote fusion [2]. The structural physiognomies of virus coats (capsids) are extremely appropriate to virus propagation. The coat must enclose and protect the nucleic acid, flexible against interference be talented of broaching the outer wall of a target cell and provide a confident pathway for attending nucleic acid into the target cell. Hollow spikes on the capsid fulfill the latter two roles, from which it has been deduced that they must have unusually high strength and stiffness in axial compression [3]. The spike protein (S protein) is a type I transmembrane protein, a sequence of amino acids ranging from 1,160 for avian infectious bronchitis virus (IBV) and up to 1,400 amino acids for feline coronavirus [4]. Spike proteins assemble to create the distinctive “corona” or crown-like look in trimers on the surface of the virion. The ectodomain of all CoV spike proteins portions the same organization in two domains: a receptor-binding N-terminal domain called S1 and a fusionresponsible C-terminal S2 domain The variable spike proteins (S proteins) reflect CoV diversity, which have developed into forms that vary in their receptor interactions and their reaction to different environmental triggers of virus-cell membrane fusion [5]. A notable peculiarity between the spike proteins of diverse coronaviruses is whether it is cleaved or not during assembly and exocytosis of virions [6]. The entry Herpesvirus and membrane fusion to the host cell equire three virion glycoproteins, gB and a gH/gL heterodimer, that function as the “core fusion machinery” [7]. The viral envelope of Ebola virus contains spikes consisting of the glycoprotein (GP) trimer. These viral spike glycoprotein docks viral particles to the cell surface, endosomal entry, and membrane fusion [8]. Acute respiratory syndrome coronavirus (SARS-CoV) is a zoonotic pathogens that traversed the species barriers to infect humans. These coronaviruses hold a surface-located spike (S) protein that recruits infection by mediating receptor-recognition and membrane fusion [9]. Bioinformatics plays an important role in all aspects of protein analysis, including sequence analysis, and evolution analysis. In sequence analysis, several bioinformatics techniques can be used to provide the sequence comparisons. In evolution analysis, we use the technique like phylogenetic trees to find homologous proteins and identified the most related taxa. With bioinformatics techniques and databases, function, structure, and evolutionary history of proteins can be easily identified [10]. Several bioinformatics methods can be used in sequence analysis to provide sequence comparisons.
Materials and Methods
Sequences, alignment, and construction of phylogenetic tree
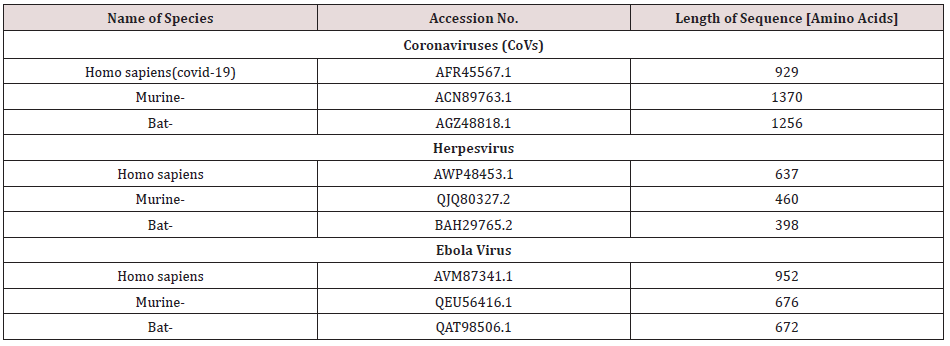
Amino acids sequences for the spike protein of Covid-19, Ebola, and herpes simplex viruses from bats, murine as well as Homo sapiens were taken from the National Center for Biotechnology Information database. The accession numbers of the corresponding database entries and species names are listed in Table 1 and Figure 1. Sequences were aligned with by CLUSTALW [11]. Selection of conserved blocks was performed using GBlocks (version 0.91 to eliminate divergent regions and poorly aligned positions using standard settings according to Castresana [12]. The Akaike information criterion (AIC) were performed, using a maximum-likelihood (ML) starting tree to estimates the quality of each model, relative to each of the other models (Table 2). The most suitable model was WAG+G+F [13]. Then phylogenetic tree was constructed with neighbor-joining and maximum likelihood algorithms using MEGA version 5 [14]. The stability of the topology of the phylogenetic tree was assessed using the bootstrap method with 100 repetitions [15].
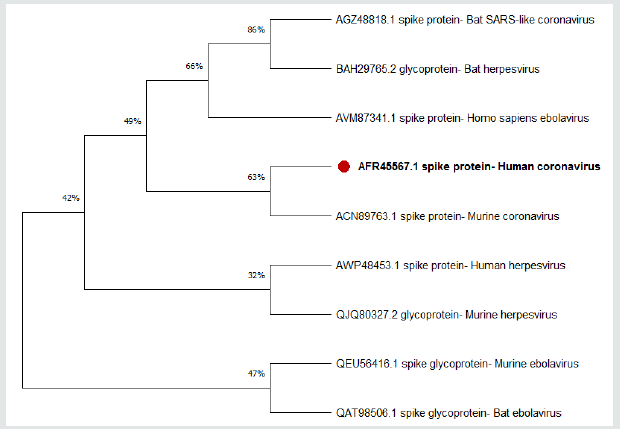
Figure 1: Multiple sequence alignment of Spike protein of coronaviruses (CoVs), Ebola, and herpes simplex viruses from human , bat and Murine.

Computation of the theoretical pI (isoelectric point) of protein sequences
Estimation of the isoelectric point (pI) based on the amino acid sequence was determined using isoelectric Point Calculator (IPC), a web service and a standalone program for the accurate estimation of protein and peptide pI using different sets of dissociation constant (pKa) values [16]. Models with the lowest BIC scores (Bayesian Information Criterion) are considered to describe the substitution pattern the best. For each model, AICc value (Akaike Information Criterion, corrected), Maximum Likelihood value (lnL), and the number of parameters (including branch lengths) are also presented [1]. Non-uniformity of evolutionary rates among sites may be modeled by using a discrete Gamma distribution (+G) with 5 rate categories and by assuming that a certain fraction of sites is evolutionarily invariable (+I). Abbreviations: TR: General Time Reversible; JTT: Jones-Taylor-Thornton; rtREV: General Reverse Transcriptase; cpREV: General Reversible Chloroplast; mtREV24: General Reversible Mitochondrial.
Results
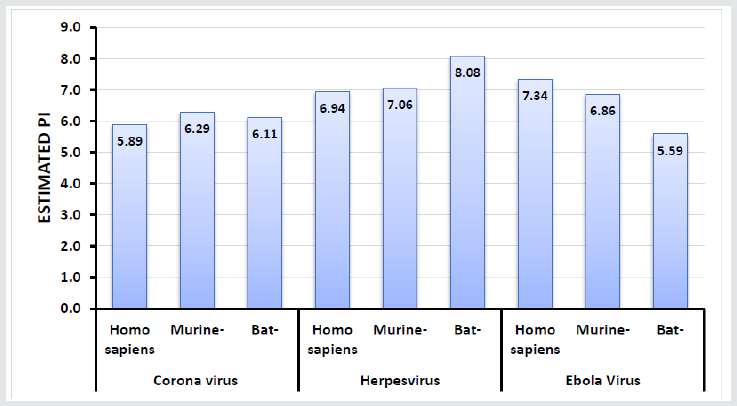
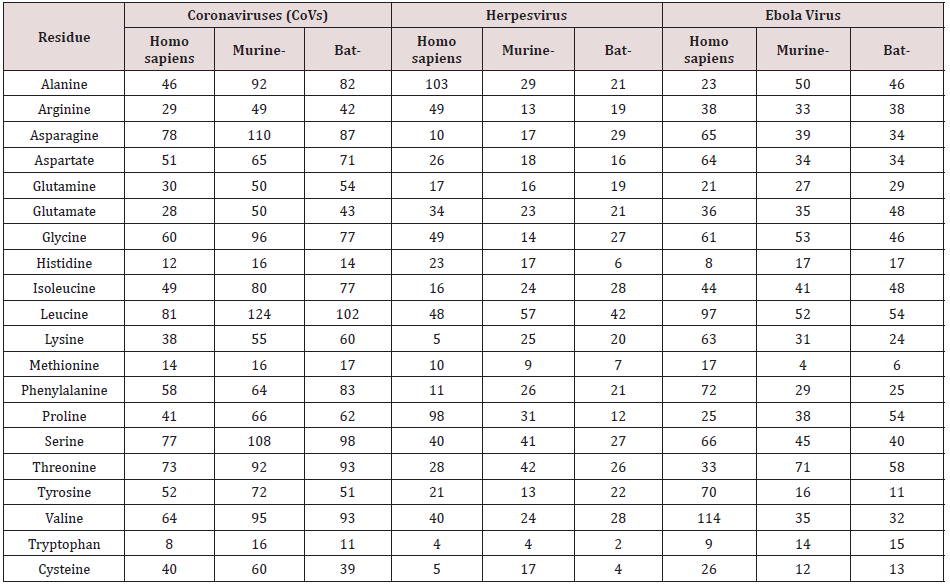
In order to unravel the phylogenetic relationship of Spike protein between the different taxa, a phylogenetic consensus tree was constructed using the Bayesian Inference (BI) and Maximum Likelihood (ML) methods (Figure 1). The present results revealed that the identity between different taxa was nonsignificant, However alignment of human Coronavirus (Covid-19) and Bat Coronavirus revealed identity equal to 57.98% followed by Murine Coronavirus which displayed 27.61% when compared by human Coronavirus. The chemical composition of the tested protein is illustrated in Table 3. Figures 2 & 3 show the correlation plots between the theoretical isoelectric points for spike protein of different corona virus in different species. The current results displayed that estimated pI of spike protein sequence in three investigated viruses in different species ranges from 5.59 to 8.08. With highest value for spike protein in herpesvirus of bat. The Estimated charge over pH range of the investigated were listed in Table 4. The current results revealed that the behavior of s protein in different species exhibited different estimated charge at different pH ranges. Some S protein reveled low negative charge as pH increases such as Ebola Virus of bat while S protein of Corona virus in murine showed high negative charge as pH elevated.
Figure 2: The phylogenetic tree shows the relationship of the Spike protein of coronaviruses (CoVs)with protein sequences from other species. The maximum likelihood tree is based on complete coding sequences.
Discussion
One aspect that may provide some insight into the interactions of the S protein is the electrostatic potential it generates. the affinity constant for the receptor-binding domain (RBD) of viron protein potentially contributing to its transmission efficiency [17]. The S protein remains predominantly in the closed conformation to mask its receptor-binding domains (RBDs), thereby impeding their binding. To bind with ACE2, the S protein transforms into its open conformation, revealing its binding interface [18]. The present study relies on a comparative investigation, regarding the identity of spike protein of 3 viruses (Covid-19, Ebola, and herpes simplex) to identify the most related taxa. Our investigation revealed high similarities of Spike protein of coronaviruses in human and bats [19]. The computed amino acid composition of spike protein. Several residues showed a significant difference between the compositions in spike proteins in the three investigated virus in different species. This result reveals the importance of specific residues in these classes of proteins. The polar uncharged residues, especially Serine, Asparagine, and Glycine, have higher occurrence in spike protein of murine, which are important for the folding, stability, and function of such class of proteins [20]. Our analysis revealed that the Coronavirus spike protein of murine may be more efficient in discovering suitable vaccine for Coronavirus. The S protein amino acids variations among different coronaviruses such as (SARS, herpes and ebola). The SARS-CoV-2 virus shares 57.98% of its genome with the other bat coronaviruses. The sequence identity in the S protein bat coronavirus appears to be the closest relative of SARS-CoV-2. Our results in accordance with Rothan [21]. The isoelectric point (pI) is the pH value at which a molecule’s net charge is zero [22]. Information about the isoelectric point is important because the solubility and electrical repulsion are lowest at the pI. Hence, the tendency to aggregation and precipitation is highest. In the case of viruses, the value thus provides information about the viral surface charge in a specific environment [23]. In polar media, such as water, viruses possess a pH-dependent surface charge [23]. This electrostatic charge governs the movement of the soft particle in an electrical field and thus manages its colloidal behavior, which plays a major role in the processes of virus entry. The pH value at which the net surface charge changes its sign is called the isoelectric point and is a characteristic parameter of the virion in equilibrium with its atmosphere in water chemistry [24]. For some viruses the attachment influences that encourage binding to accommodating cells are extremely definite, but the arrangement of actions that activates viral entry is only now establishment to be understood. The charge of attachment protein may play an important role in attachment and entry of virus [25]. The current results revealed that the pH affect the net charge of S protein of different taxa with different behavior. All the investigated taxa exhibit increases in negative charge as the pH increased except for the Ebola virus form Bats which showed unstable behavior regarding the S protein charge.
Conclusion
In our study, we have investigated the variation of pHdependent changes in charges of a protein. The current results revealed that the pH affect the net charge of S protein of different taxa with different behavior.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gelderblom HR (1996) Structure and classification of viruses. In: Medical Microbiology 4th University of Texas Medical Branch at Galveston, USA.
- Arvin A, Campadelli Fiume G, Mocarski E, Moore PS, Roizman B, et al. (2007) Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, UK.
- Feughelman M (2002) Natural protein fibers. Journal of Applied Polymer Science 83(3): 489-507.
- Belouzard S, Millet JK, Licitra BN, Whittaker GR (2012) Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4(6): 1011-1033.
- Li F (2016) Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3(1): 237-261.
- Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. In: Coronaviruses. Springer p. 1-23.
- Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, et al. (2012) Herpes virus fusion and entry: a story with many characters. Viruses 4(5): 800-832.
- Beniac DR, Booth TF (2017) Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å Sci Rep 7: 46374.
- Lu G, Wang Q, Gao GF (2015) Bat-to-human: spike features determining ‘host jump’of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol 23(8): 468-478.
- Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5(4):276-287.
- Thompson JD, Gibson TJ, Higgins DG (2003) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinforma 2: 3.
- Castresana J (7000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4): 540-552.
- Wood SN (2011) Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. Journal of the Royal Statistical Society Series B (Statistical Methodology 73(1): 3-36.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10): 2731-2739.
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (NY) 39(4): 783-791.
- Kozlowski LP (2016) IPC--isoelectric point calculator. Biol Direct 11(1): 1-16.
- Wang Y, Liu M, Gao J (2020) Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci 117(25): 13967-13974.
- Zimmerman MI, Porter JR, Ward MD, Singh S, Vithani N, et al. (2020) Citizen scientists create an exascale computer to combat COVID-19. BioRxiv.
- Lau SKP, Li KSM, Tsang AKL, Lam CSF, Ahmed S, et al. (2013) Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory sy. J Virol 87(15): 8638-8650.
- Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions. Bioinforma Genet 317: 280-289.
- Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 109: 102433.
- Kozlowski LP (2017) Proteome-pI: proteome isoelectric point database. Nucleic Acids Res 45(D1): D1112-D1116.
- Michen B, Graule T (2020) Isoelectric points of viruses. J Appl Microbiol 109(2): 388-397.
- Langlet J, Gaboriaud F, Gantzer C, Duval JFL (2008) Impact of chemical and structural anisotropy on the electrophoretic mobility of spherical soft multilayer particles: the case of bacteriophage MS2. Biophys J 94(8): 3293-3312.
- Connell BJ, Lortat-Jacob H (2013) Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front Immunol 4: 385.