Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2641-1709
Research Article(ISSN: 2641-1709) 
Oral Microbiome of Adults in Sound and Cariogenic Biofilm with and without Dental Caries. A Pilot Study Volume 8 - Issue 2
Moncada G1*, Ahumada D2, Arévalo V2, De Calisto J3, Moncada X4 and Gonzalez S2
- 1Department of Oral Rehabilitation, Universidad de Los Andes, Dental School, Chile
- 2Universidad Mayor, Dental School, Faculty of Science, Chile
- 3Universidad Mayor, Biotechnology School, Faculty of Science, Chile
- 4Universidad de Chile, Molecular Biology Laboratory, Faculty of Chemical and Pharmaceutical Sciences, Chile
Received: April 13, 2022; Published: April 20, 2022
Corresponding author: Gustavo Moncada, Department of Oral Rehabilitation, Universidad de Los Andes Dental School, Santiago, Chile
DOI: 10.32474/SJO.2022.08.000282
Abstract
Aim: The objective of the present study was to identify and compare the microbiomes of healthy and caries disease in dental surface from Chilean adults.
Materials and Methods: A total of 56 site-specific supragingival biofilm samples were collected from 14 patients recruited of both sexes (18-35 years old); of them, n=5 was caries free and n=9 was caries active (ICDAS 1-3). Biofilms were analyzed by 16S ribosomal RNA amplicon sequencing. Sample microbiotas were compared at Genus and Family levels.
Results: Proportion of Streptococcus decreased in healthy patients while Veillonella increased in caries-active patients. Streptococcus population decreasing was related to the increasing of Corynebacterium, Neisseria, Leptotrichia, Capnocitophaga, Provetella and Campilobacter in healthy patients. The increasing of Streptococcus was associated with the rise of Lautropia, Haemophilus, Bengeyella and Aggregabacter in caries-active patients. In general, men had significantly higher levels of Lachnospiraceae, Aerococcaceae and Leptotrichiaceae than female.
Conclusions: There were no typical bacteria families or genera only in healthy or only caries-active patients. Caries-free and cariesactive tooth surfaces share similar microbiomes but in different proportions. Greater relative abundance of bacterial families and genera were observed in caries free dental biofilms. The bacterial diversity was lower in the caries-active group, suggesting that the bacterial diversity establishes a protective environment.
Keywords: Caries; bacteria; metagenome; microbiome; dental biofilm; 16S rRNA
Introduction
Dental caries is the most common non transmissible disease of the human being, both in developed and underdeveloped countries, and leads the group of the four chronic diseases with the highest cost of treatment [1]. In Chile, almost 100% of the adult population has experienced dental caries disease [2], similar to what has been observed in another country, with special vulnerability in the low and middle-low socioeconomic levels [3]. According to the most updated data, 35% of the population worldwide has untreated caries lesions and in Latin America this number is close to 45% [1]. Caries strongly affects quality of life, especially self-esteem and relationship life, through the impact they have on chewing, function and aesthetics [4–6]. The human oral microbiota, the consortium of the microbial inhabitants in the mouth, has been progressively recognized as playing a major role in the maintenance, promotion and alteration of health, establishing that the center of the discussion is the stability of the microbial population in the healthy mouth as a basis for understanding how these populations are transformed into a dysbiotic state that lead to caries disease [7,8]. The etiology of dental caries remains unclear because of our limited understanding of the complex oral microbiomes [9], especially after it was detected that a great number of the oral bacteria have never been cultivated [10]. Traditional studies, based on microbiological cultures established that mutans streptococci (MS) and lactobacilli (L) were the main causal agents of dental caries [9]. However, other bacteria had been detected later as participating in the dental caries process, in which acid-creating bacteria are the vehicle to penetrate enamel and allow dentin degrading microorganisms to expand the lesion. Microbial composition at the initial, enamelaffecting stage of caries is significantly different from that found at subsequent stages, as well as from dental plaque of sound tooth surfaces, the relative amount of MS increases from 0.12% in healthy enamel biofilm, to 0.72% in enamel caries [10]. During the last years other microbial species besides MS have been isolated and related to the disease such as Scardovia wiggsiae, Parascardovia denticolens, as well as several Veillonella, Prevotella Atopobtum and Propionibacterium species [10–14] and related to caries free such as Streptococcus dentisani, S. sanguinis, Actinomyces naeslundii, Rothia aeria, Neisseries [9,15,16], Streptococcus A12 [17] and AS14 [18]. Those findings have been possible thanks to the application of pyrosequencing to PCR materials of the 16S rDNA gene, which has become an exceptionally powerful approach, combined with bioinformatics processes that have simplified the detection and revealed that caries are particularly diverse ecological systems [15,19]. Next-generation sequencing (NGS) technology has exposed the complexity of oral microbiomes and provided a basis to understand how hundreds of bacterial species coexist and functionally interrelate to maintain homeostasis and give rise to health or disease conditions [7,20,21].
Using culture-dependent and independent methods, it is estimated that the diversity of the oral bacteria includes >700 different microbial species [14,22,23]. Later studies using NGS technologies to analyze the species of the oral cavity suggest this is constituted by over 19,000 phylotypes [24]. Despite these enormous advances in the bacterial caries community, they are insufficient to understand the onset and progress of the disease. Rapid and accurate identification of oral microbiota imbalance could be critical to provide the appropriate treatment for patients suffering caries disease. Traditional clinical microbiology methods are less able to detect potential oral microbiota imbalance, but recent advances in amplification and NGS, in particular applied to the bacterial ribosomal RNA encoding genes (16S rRNA genes) have overcome this problem, are increasingly used in the microbiology analysis [25] and have enormously expanded our knowledge of microbiome composition [26]. There are numerous caries risk assessment (CRA) models, promoted for disease management and for identification of individuals with risk of future caries. CRA is considered essential for the planning of preventive and therapeutic strategies, but the scientific evidence relating to standardized CRA models is still limited, no sufficient evidence is available to support which method is the most effective model in CRA and prediction, and standardized CRA models still remains limited [27]. Traditionally, the risk index for dental caries has been the previous clinical caries experience, diet, dental hygiene, heredity, immunity and diseases of the patient. While bacteria are the main etiological factor of carious lesions, the CRA does not consider bacterial quantification or focus solely on the level of S. mutans in saliva. At present, with the advent of massive molecular techniques, a possibility for addressing this issue is to study the oral microbiome [28]. The objective of the present study was to identify and compare the microbiomes of sound and cariogenic dental biofilm in adults, to know the differences between both microbiomes that allow us to enrich the analysis of patients’ cariogenic risk.
Material and Methods
Subjects, processing and sequencing: 14 individuals (7 female and 7 male) between 18 - 35 years old (x ̅=24.7), with the presence of 28 teeth in the mouth (excluding third molars), individuals ICDAS=0, and individuals with caries lesions classified as ICDAS 1-2-3, without periodontal or systemic diseases were included. The exclusion criteria were intake of any medication, treated with antibiotics or fungicides at least 1 month before taking the sample, use of oral antiseptics or toothpaste with arginine, or disagreement with the informed consent. The evaluation of caries lesion severity was carried out by International Caries Detection and Assessment System criteria (ICDAS) [29]. The cariogenic risk assessment was made according to CAMBRA criteria (30) and a daily diet questionnaire was requested, during the last week. The informed consent was achieved under a protocol approved by the Institutional Research Board at Universidad Mayor (302017). At baseline, patients were clinically grouped by caries status as Caries Free (CF), and patients with active caries lesions (AC) (ICDAS 1-3). Recruits did not perform brushing or any oral hygiene procedure for at least 24 hours before sampling. In addition, supragingival biofilm samples were taken at least 8 hours after fasting.
Sample collection and 16S rRNA gene sequencing: Supragingival dental biofilm samples were collected from the enamel of the mesiobuccal surface of the first 4 first molars, without direct contact with the caries lesion or gingival tissue, by periodontal curettes which were transferred to a buffer solution. Each sample was transferred into a vial containing a lysis and stabilization buffer that preserves the rRNA for transport at ambient temperatures. Samples were lysed using bead-beating, and rRNA was extracted in a class 1000 clean room by a guanidine thiocyanate silica column-based purification method using a liquid-handling robot [31,32]. PCR amplification of the 16S rRNA genes was performed with universal primers amplifying the V4 variable region (515F: GTGCCAGCMGCCGCGGTAA and 806R: GGACTACHVGGGTWTCTAAT) [33], containing Illumina tags and barcodes. Samples were barcoded with a unique combination of forward and reverse indexes allowing for simultaneous processing of multiple samples. PCR products were pooled, column-purified, and size-selected through microfluidic rRNA fractionation [34]. Consolidated libraries were calculated by quantitative real time PCR using the Kapa Bio-Rad iCycler qPCR kit on a BioRad MyiQ previously loading into the sequencer. Sequencing was performed in a pair-end mode on the IIlumina NextSeq 500 platform rendering 2 x 150 bp pairend sequences. All samples pass the 10000 reads quality control threshold. We used the threshold to ensure detection of all targeted taxa, even at low abundance.
Taxonomic annotation and reference database generation: After sequencing, demultiplexing of samples was made using Illumina´s BCL2FASTQ algorithm. Reads were filtered using an average Q-score >30. Forward and reverse reads were appended together after exclusion of primers and any leading bases, and clustered using version 2.1.5 of the Swarm algorithm [35] using a distance of one nucleotide and the “fastidious” and “usearch-abundance” flags. The most abundant sequence per cluster was considered the real biological sequence and was assigned the count of all reads in the cluster. The residual reads in a cluster were considered to contain errors as a product of sequencing. The representative reads from all clusters were subjected to chimera exclusion using the VSEARCH algorithm [36]. Reads passing all above filters (filtered reads) were aligned using 100% identity over 100% of the length against a hand-curated database of target 16S rRNA gene sequences and taxonomic annotations derived from version 123 of the SILVA database [37]. The hand-curated databases for each taxon were created by selectively eliminating sequences with amplicons that were ambiguously annotated to more than one taxonomic group. The most predominant bacterial families were recognized in both groups (AC-CF). The relative abundance of each taxon was determined by dividing the count linked to that taxon by the total number of filtered reads.
Statistical analysis
To determine the normality of the data the Shapiro-Wilk test was used. To compare the oral microbiome of patients with and without caries and men with women the unpaired Student test was performed for the majority of data. However, five families (Burkholderiaceae, Cardiobacteriaceae, Propionibacteriaceae, Pseudomonadaceae and Fusobacteriaceae) and three bacterial genera (Lautropia, Bergeyella and Kingella) were out of the normality; therefore, the Mann-Whitney U test was performed. Statistical software Stata® 15.1 and a level of statistical significance with a value of p≤0.05 were used.
Results
Families’ relative abundance analysis
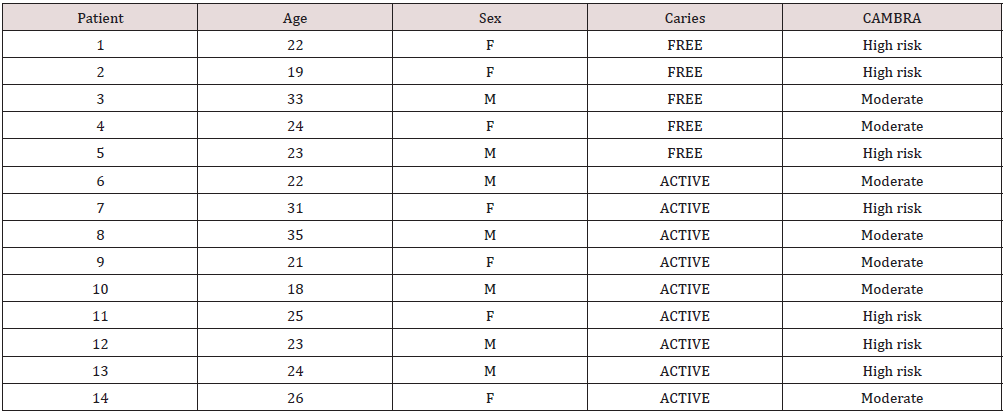
A total of 56 site-specific supragingival biofilm samples were collected, grouped in 5 caries-free patients and 9 with 1 or more active caries lesions (ICDAS 1-3), (Table 1). Individuals affected by active caries (n=9) and caries-free (n=5) were compared by their oral bacterial microbiomes. On average, 25 families were identified in the group affected by caries, and 26 in the cariesfree group. The 20 most abundant bacterial families were analyzed, to distinguish them from those with lower relevance percentages (Figure 1A). In the 20 most abundant bacterial families, it was found that the group of individuals with caries had greater abundance of Streptococcaceae, Pasteurellaceae, Veillonellaceae and Corynebacteriaceae. By contrast, in caries free individuals, Veillonellaceae, Corynebacteriaceae, Neisseriaceae and Streptococcaceae were more predominant, in decreasing order (Figure 1B). The only family that presented significant difference was Prevotellaceae (p=0.017). The three lesser relative abundance bacterial families found for both groups of patients were the same. These families were Mycobacteriaceae, Peptostreptococcaceae and, Peptococceae. In the CF group these families showed a higher relative abundance than AC group. (non-significant).
Genera relative abundance analysis
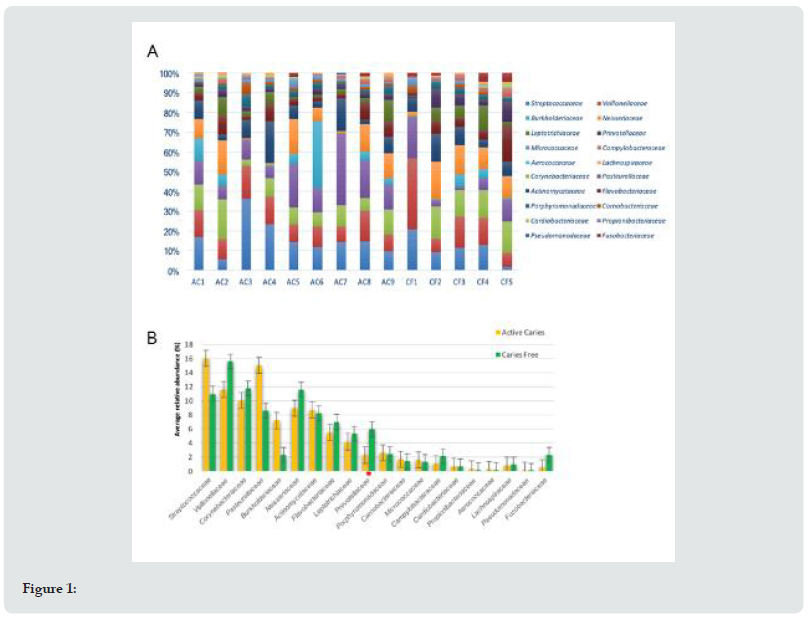
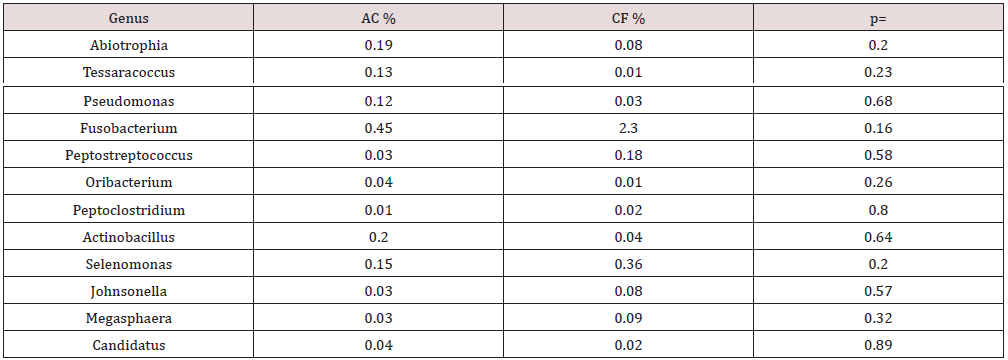
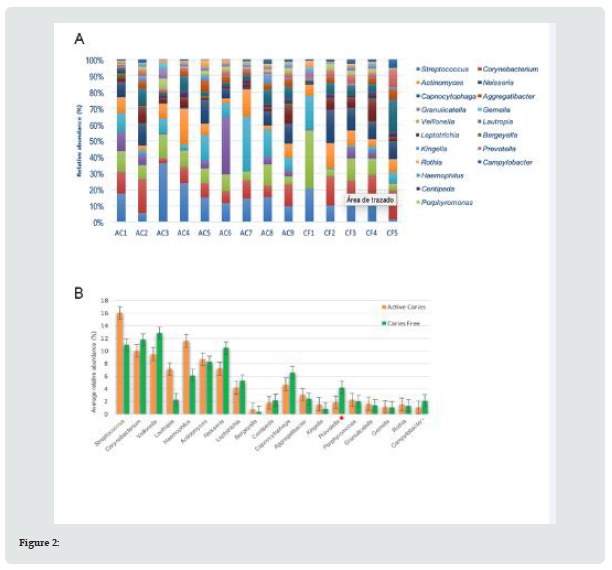
On average, 43 bacterial genera were found in the caries group (AC), and 47 genera in the caries-free group (CF). For both groups, 19 bacterial genera were considered the most abundant (Figure 2A). It was found that the AC group had greater abundance of Streptococcus, Haemophilus, Corynebacterium and Veillonella. By contrast, in the CF group, Veillonella, Corynebacterium, Streptococcus and Neisseria were more predominant, in decreasing order. Significant differences were only observed in the Prevotella genus level (p=0.005) (Figure 2B). The same 10 genera showed lesser relative abundance in both groups (AC and CF). Interestingly, in 6 genera a significant higher relative abundance was observed in the CF group compared with the AC group (Table 2).
Families and Genera analysis related to CAMBRA caries risk
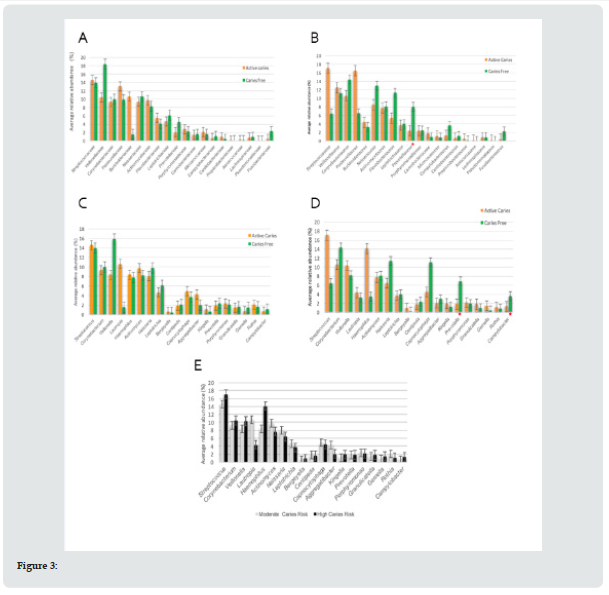
The dietary diaries of all subjects were analyzed for seven days, with no relevant differences in both groups, when analyzing the type of food consumed and daily intake frequency. In moderate risk patient’s similar amounts of bacterial families were observed in AC (n=25) and CF groups (n=26). In high-risk individuals, in AC group n=24 families, versus n=28 families in CF group. In patients with moderate risk, it was found that the AC group had greater abundance of Streptococcaceae, Pasteurellaceae, Burkholderiaceae and Veillonellaceae in decreasing order. The CF group had greater abundance of Veillonellaceae, Streptococcaceae, Neisseriaceae and Corynebacteriaceae, in decreasing order (Figure 3A). In patients with high cariogenic risk, the highest abundance was for Streptococcaceae, Pasteurellaceae, Veillonellaceae and Corynebacteriaceae. For those without caries disease, Corynebacteriaceae, Neisseriaceae, Flavobacteriaceae and Veillonellaceae were more abundant (Figure 3B). Only for Prevotellaceae family were significant differences found (p=0.017).
At the genera level, in high-risk individuals, 49 different genera were observed in the CF group, and 42 genera in the AC group. Similar results were observed in individuals of moderate risk. In the CF group more bacterial genera were found (n=45) than in the AC individuals (n=44). The moderate-risk AC individuals had greater abundance of Streptococcus, followed by Lautropia, Actinomyces and Corynebacterium. Meanwhile, in CF individuals, Veillonella predominated, followed by Streptococcus, Corynebacterium and Neisseria. In the high risk group, AC individuals showed greater abundance of Streptococcus, Haemophilus, Corynebacterium and Veillonella. By contrast, among the caries-free, Corynebacterium, Neisseria, Capnocytophaga and Veillonella predominated (Figures 3C & 3D). Patients with high caries risk and clinically CF, presented a significantly higher abundance of the bacterial genera Prevotella (p=0.05) and Campylobacter (p=0.05). Subsequently, we compared relative abundance at the families and genera levels, between high and moderate risk with active caries. High caries risk patients presented higher family abundance of Streptococcaceae, Pasteurellaceae, Veillonellaceae and Corynebacteriaceae, while for moderate caries risk Streptococcaceae, Pasteurellaceae, Burkholderiaceae and Veillonellaceae predominated. Among the high risk the genera Streptococcus, Haemophilus, Corynebacterium and Veillonella in greater abundance and for moderate risk, Streptococcus, Lautropia, Actinomyces and Corynebacterium (Figure 3E).
Relative abundance differences between sexes
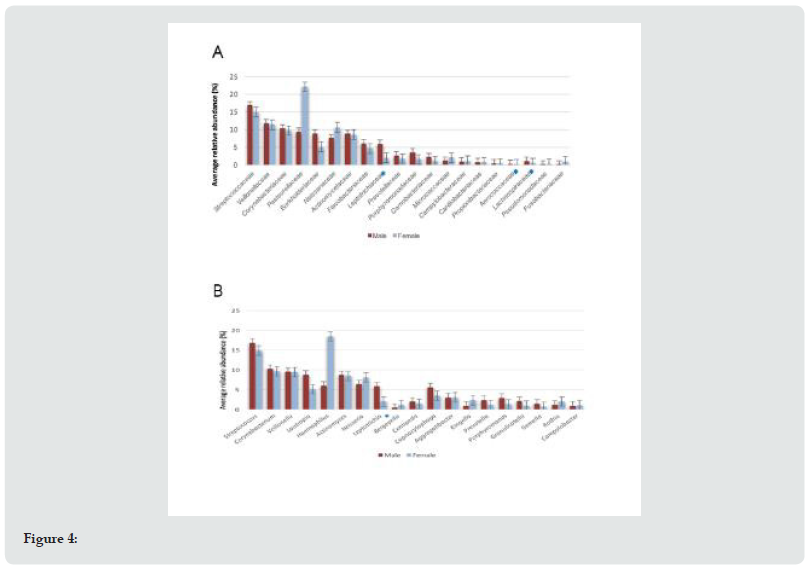
Men and women presented the same average number of bacterial families (n=25). While in bacterial genus it was different, men (n=45) compared to women (n=40). A significantly higher abundance at the family level was observed in men, with families Leptotrichiaceae (p=0.04), Aerococcaceae (p=0.04) and Lachnospiraceae (p=0.03), and a significantly greater abundance only in the genus Leptotrichia (p=0.04) (Figures 4A & 4B). Men showed greater relative abundance of the bacterial families Streptococcaceae, Veillonellaceae, Corynebacteriaceae and Pasteurellaceae. In the case of women, families Pasteurellaceae, Streptococcaceae, Veillonellaceae and Neisseriaceae were more abundant, in descending order. In bacterial genus, it was found that men had more relative abundance of Streptococcus, Corynebacterium, Veillonella and Lautropia. For women it was Haemophilus, Streptococcus, Corynebacterium and Veillonella, in that order.
Discussion
The ecological balance in the oral cavity is maintained, in healthy conditions, through interactions between the various microorganisms present, through symbiotic and/or antagonistic processes. The imbalance between the members of the oral microbiota can generate changes in the environment through various metabolic activities, facilitating the development of diseases such as dental caries [38]. For this reason, it is of great interest to understand the dynamic behavior of the oral microbiomes in health and disease. In this pilot study, the abundance and diversity of families and bacterial genus from 14 adults with and without caries disease were analyzed and compared. Significant differences of greater abundance of Prevotellaceae and Prevotella were found in healthy individuals. Additionally, a trend of dominance was observed for the families Streptococcaceae, Pasteurellaceae, Burkholderiaceae, and the genera Streptococcus, Lautropia, Haemophilus, in the Active Caries group in comparison to the Caries Free group. The greater capacity of aggregation and coaggregation that Streptococcus has in comparison to other bacteria, would explain why they tend to be one of the most predominant microorganisms in this and in other studies about the oral microbiome, since they allow the beginning of the biofilm formation process [39]. However, some species of Streptococcus would be associated with stages of health or disease [40]. Interestingly, while the proportion of Streptococcus was lower in healthy patients, Veillonella increased in CF patients, similar to the conclusions of Mashima et al. [41]. Also, the Streptococcus decreasing was related to the increase of Corynebacterium, Neisseria, Leptotrichia, Capnocitophaga, Prevotella and Campylobacter in healthy patients, similar to observations by Lee et al. [42].
Within the family Pasteurellaceae, the genera Haemophilus and Actinobacillus were in greater abundance in individuals with caries. In agreement with the present study, greater proportions of Haemophilus have been identified in saliva of patients with caries [43] and in non-cavitated enamel lesions [7]. Haemophilus species have the capacity to reduce nitrate to nitrite and produce lactic, succinic and acetic acid when fermenting glucose [44,45]. It is known that nitrite inhibits the production of acid by other bacteria, which would be a protective factor for caries. It is therefore necessary to identify which species and which metabolic activities predominate, and in what circumstances. Other bacteria that predominated in CF individuals compared to the AC group, such as Capnocytophaga, Campylobacter, Peptostreptococcus and Fusobacterium, have been described in other reports as contributors to oral health [46,47,51– 53]. Edlund et al. showed that Fusobacterium was highly active at pH 7 and presented evident decrease at lower pH [49], and can also produce ammonium [7], which has been associated with the ability to significantly increase the pH in the biofilm and can inhibit the action of the potentially cariogenic microbiota [50]. As mentioned, Prevotella was most abundant in CF versus AC group. Yang et al. [51], described that overpopulation of Prevotella has been observed in active caries, but this observation was made in saliva, while the current study was made in dental biofilm. This could be due to the fact that for certain Prevotella species commonly found in the oral cavity, obtaining energy depends mainly on the metabolism of nitrogen compounds, producing ammonium [52]. However, they can also metabolize glucose, lowering the pH. Thus, in stages of health where the pH is usually higher, the metabolization of nitrogen compounds predominates. In late stages, there could be a predominance of glucose metabolism instead, increasing its rate of growth, which would explain the higher levels of Prevotella in dentinal lesions found in various studies [48,53]. Furthermore, as other reports suggest, the various Prevotella species are not distributed in a similar way between the two groups of individuals (AC-CF), therefore, the resolution at the species level is necessary [54].
The bacterial diversity was lower for the group of individuals with caries, in the two taxonomic levels studied. This agrees with previous studies [53,55–57], justifying that the genes associated with the sugar-fermenting capacity and acid tolerance of acidogenic microorganisms would be overrepresented in incipient caries, generating a decrease in the pH of the medium and allowing a more selective habitat by leading to the suppression of acid-sensitive bacteria [58]. As a result, despite the fact that both groups presented bacteria with sugar-fermenting capacities, finding less bacterial diversity in the group of AC individuals is related to the increase in selectivity of the mentioned medium [58]. When we compared the oral microbiome in individuals with and without caries within the moderate and high cariogenic risk groups, it was found that for high risk, Prevotellaceae, Prevotella, and Campylobacter genera were significantly higher in healthy individuals. This, along with the trend observed in the rest of the results, was similar to what was obtained when comparing the oral microbiome between individuals with and without caries not categorized in a certain risk, suggesting that the differences in the oral microbiome are directly related with the presence or absence of the disease rather than in the cariogenic risk CAMBRA assessment.
In fact, independent of the risk, Prevotellaceae, Prevotella, Campylobacter, Neisseria and Fusobacterium have marked the trend to be more noticeably present in CF individuals, while Streptococcaceae, Streptococcus, Pasteurellaceae, Haemophilus, Actinobacillus and Oribacterium predominated in AC individuals. Furthermore, given the results obtained when analyzing the oral microbiome only in individuals with caries according to their cariogenic risk, we propose that this does not primarily influence the abundance of one bacterium over another, but rather that bacterial diversity establishes the protective environment. In addition, automatically categorizing a subject as “high cariogenic risk” due *to presenting dental restorations performed in the last three years may be inadequate if we consider evidence that the caries risk level can be reduced with behavioral change and therapeutic intervention within a period of 18 to 24 months [59].
On the other hand, non-relevant difference findings in the type of food consumed and in the frequency of daily intakes among groups of individuals with and without caries, confirms the need to have a global view of the disease, since although frequent intake of sugars is a transcendental factor, it only constitutes one of the many pieces of the puzzle. In the present study, the non-difference of diet factor between groups could be explained by the low sample size. Finally, several studies have compared the prevalence of caries disease in men and women, placing women as the most atrisk [60–62]. In the current study, men with active caries showed significant higher levels of Lachnospiraceae, Aerococcaceae and Leptotrichiaceae. It has been mentioned that Leptotrichiaceae (Leptotrichia) has an evident cariogenic potential [63]. On the other hand, Fusobacterium, described as asaccharolytic or weakly fermentative [7], was the only genus found significantly more in women. Prevotella tended to be found more in males, despite not having significant differences with females. By contrast Haemophilus tends to be found higher in females. In general, there would be a subtle difference between both sexes. In men, the more predominant have mostly acidogenic capacities, while in women, although bacteria are capable of generating acidic products, they also have metabolic pathways capable of exerting a regulating role in pH, like the production of ammonium by Fusobacterium and Prevotella, and nitrite by Haemophilus. However, we do not know what metabolic activity predominates in these. It is striking that although the number of bacterial families was the same for both groups of individuals, there was a lower genera abundance in women. This would be interesting to investigate, since as mentioned previously, from a microbiological point of view, a more diverse community represents a more stable ecosystem.
In the present study, the sampling was performed supragingivally, on the mesiovestibular surfaces of the first molars, without direct contact with the carious lesion. In relation to this, there is evidence that establishes that bacterial diversity is greater at molars’ proximal and lingual surfaces, and that occlusal and proximal surfaces would be more susceptible to developing lesions of caries, harboring more acidogenic species [64]. On the other hand, it has been pointed out that at the level of incipient carious lesions, the microbial component tends to increase its acidogenic capacity and decrease its diversity with respect to healthy areas [55]. In our study, the samples were taken only in healthy areas; therefore, it would be interesting to contrast it with samples obtained directly from the carious lesion, to check if this situation manifests itself more clearly. Additionally, it should be emphasized that the present study was carried out through the sequencing of the 16S rRNA gene, which provides a quantitative description of the bacteria present in an ecological niche [59]. However, it does not allow us to safely understand the degree of cariogenicity of the different oral microorganisms since these are highly heterogeneous, suggesting to the authors that this method allows the knowledge of only one of the variables involved for a more indepth understanding of the behavior of the caries disease. This pilot study is an approach for understanding the oral microbiome in stages of health and caries disease in adult individuals. It also provides a look at how sex or cariogenic risk could be related to the disease and contributing to the knowledge of barely studied taxa. Nevertheless, although certain biochemical processes are known to some extent to be involved with various bacteria in the oral cavity, it cannot be established which microorganisms are active, nor which metabolic pathways are carried out in certain situations. As a result of this, future metatranscriptomic or metabolomic approaches should be considered, which would allow identifying key molecules that could interfere with the onset and/or progression of caries, contemplating the species level. The latter is necessary because it is supposed that differences would be noticeably widened and could provide more accurate information about caries microbiomes. However, it is well known that in vivo conditions are practically impossible to control, considering the availability of nutrients in the environment, high interpersonal variability and the heterogeneous spatial distribution of microorganisms, among others [7], so the study of oral microbiota clearly continues to be a great challenge.
Conclusion
There were no typical bacteria families or genera only in healthy or only caries-active patients. Caries-free and caries-active tooth surfaces share similar microbiomes but in different proportions. Greater relative abundance of bacterial families and genera were observed in caries free dental biofilms. The bacterial diversity was lower in the caries-active group, suggesting that the bacterial diversity establishes a protective environment.
Declaration of Interest
None. This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
References
- Kassebaum NJ, Bernabé E, Dahiya M (2015) Global Burden of Untreated Caries: A Systematic Review and Metaregression. J Dent Res 94: 650-658.
- Urzua I, Mendoza C, Arteaga O (2012) Dental caries prevalence and tooth loss in chilean adult population: first national dental examination survey. Int J Dent 2012: 810170.
- (2003) Ministério da Saúde, Ministério da Saúde, Coordenação nacional de saúde bucal, condições se saúde bucal da população Brasileira 2002-2003. Resultados principais 2003.
- León S, Bravo Cavicchioli D, Correa Beltrán G, Giacaman RA (2014) Validation of the Spanish version of the Oral Health Impact Profile (OHIP-14Sp) in elderly Chileans. BMC Oral Health 14: 95.
- Sheiham A (2005) Oral health, general health and quality of life. Bull World Health Organ 83: 644.
- Martins Júnior PA, Vieira Andrade RG (2013) Impact of early childhood caries on the oral health-related quality of life of preschool children and their parents. Caries Res. 47: 211-218.
- Simon Soro A, Mira A (2015) Solving the etiology of dental caries. Trends Microbiol 23: 76-82.
- Mira A, Simon Soro A, Curtis MA (2017) Role of microbial communities in the pathogenesis of periodontal diseases and caries. Clin Periodontol 44: 23-38.
- Richards VP, Alvarez AJ, Luce AR (2017) Microbiomes of Site-Specific Dental Plaques from Children with Different Caries Status. Infect Immun 85: e00106-17.
- Simón Soro A, Belda Ferre P, Cabrera-Rubio R (2013) A tissue-dependent hypothesis of dental caries. Caries Res 47: 591-600.
- Badet C, Thebaud NB (2008) Ecology of lactobacilli in the oral cavity: a review of literature. Open Microbiol J 2: 38-48.
- Mantzourani M, Fenlon M, Beighton D (2009) Association between bifidobacteriaceae and the clinical severity of root caries lesions. Oral Microbiol Immunol 24: 32-37.
- Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R (2011) The oral metagenome in health and disease. ISME J 6: 46-56.
- Aas JA, Griffen AL, Dardis SR (2008) Bacteria of dental caries in primary and permanenet teeth in children and young adult. J Clin Microbiol 46: 1407-1417.
- Tanner AC, Kressirer CA, Faller LL (2016) Understanding caries from the oral microbiome perspective. J Calif Dent Assoc 44: 437-446.
- Camelo Castillo A, Benítez Páez A, Belda Ferre P (2014) Streptococcus dentisani sp. nov., a novel member of the mitis group. Int J Syst Evol Microbiol 64: 60-65.
- Huang X, Palmer SR, Ahn SJ (2016) A highly arginolytic streptococcus species that potently antagonizes Streptococcus mutans. Appl Env Microbiol 82: 2187-2201.
- Al Hebshi NN, Baraniya D, Chen T, Hill J, Puri S, et al. (2018) Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children. J Oral Microbiol 11: 1557986.
- Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL (2012) Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7: e47722.
- Duran Pinedo AE, Frias Lopez J (2015) Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes Infect 17: 505-516.
- Burne RA, Zeng L, Ahn SJ, Palmer SR, Liu Y, Lefebure T, Stanhope MJ, Nascimento MM (2012) Progress dissecting the oral microbiome in caries and health. Adv Dent Res 24: 77-80.
- Haffajee AD, Socransky SS, Patel MR, Song X (2008) Microbial complexes in supragingival plaque. Oral Microbiol Immunol 23: 196-205.
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr (1998) Microbial complexes in subgingival plaque. J Clin Periodontol 25: 134-144.
- Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, et al. (2008) Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87: 1016-1020.
- Woo PC, Lau SK, Teng JL, Tse H, Yuen KY (2008) Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 14: 908-934.
- Weinstock GM (2012) Genomic approaches to studying the human microbiota. Nature 489: 250-256.
- Cagetti MG, Bontà G, Cocco F, Lingstrom P, Strohmenger L, et al. (2018) Are standardized caries risk assessment models effective in assessing actual caries status and future caries increment? A systematic review. BMC Oral Health 18: 123.
- Inquimbert C, Bourgeois D, Giraudeau N, Tramini P, Viennot S, et al. (2019) Microbiota of interdental space of adolescents according to Risk of Caries: A cross-sectional study protocol. Contemp Clin Trials Commun 16: 100444.
- Ismail AI, Sohn W, Tellez M, Amaya A, Sen A, et al. (2007) The international caries detection and assessment system (ICDAS): An integrated system for measuring dental caries. Community Dent Oral Epidemiol 35: 170-178.
- Featherstone JD, Chaffee BW (2018) The Evidence for Caries Management by Risk Assessment (CAMBRA®) Adv Dent Res 29: 9-14.
- Hummel W, Kula MR (1989) Simple method for small-scale disruption of bacteria and yeasts. Microbiol Methods 9: 201-209.
- Cady NC, Stelick S, Batt CA (2003) Nucleic acid purification using microfabricated silicon structures. Biosens Bioelectron 19: 59-66.
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci UA 108: 4516-4522.
- Abdel R, Minalla AR, Dubrow R, Bousse LJ (2001) Feasibility of high-resolution oligonucleotide separation on a microchip. Proc. SPIE 4560, Microfluidics and BioMEMS pp. 90-97.
- Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M (2014) Swarm: robust and fast clustering method for amplicon-based studies. Peer J 2: e593.
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. Peer J 4: e2584.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: 590-596.
- Moon Jh, Lee JH (2016) Probing the diversity of healthy oral microbiome with bioinformatics approaches. BMB Rep 49: 662-670.
- Ofek I, Doyle RJ (2003) Bacterial Adhesion to Cells and Tissues. 1st Chapman & Hall, New York, USA pp. 54-93.
- Hoceini A, Khelil NK, Ben-Yelles I, Mesli A (2016) Caries-related factors and bacterial composition of supragingival plaques in caries free and caries active Algerian adults. Asian Pac J Trop Biomed 6: 720-726.
- Mashima I, Theodorea CF, Thaweboon B, Thaweboon S, Scannapieco FA, et al. (2017) Exploring the salivary microbiome of children stratified by the oral hygiene index. PLoS One 12: e0185274.
- Lee SE, Nam OH, Lee HS, Choi SC (2016) Diversity and homogeneity of oral microbiota in healthy Korean pre-school children using pyrosequencing. Acta Odontol Scand 74: 335-336.
- Belstrøm D, Holmstrup P, Fiehn NE, Kirkby N, Kokaras A, et al. (2017) Salivary microbiota in individuals with different levels of caries experience. J Oral Microbiol 9: 1270614.
- Haemophilus Hardy diagnostics.
- Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, et al. (2009) Bergey’s Manual of Systematic Bacteriology. 2nd ed. Springer Science & Business Media, New York, USA.
- Chen C, Hemme C, Beleno J, Shi ZJ, Ning D, et al. (2018) Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J 12: 1210-1224.
- Riggio MP, Lennon A (2003) Identification of oral peptostreptococcus isolates by PCR-restriction fragment length polymorphism analysis of 16S rRNA genes. J Clin Microbiol 4: 4475-4479.
- Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, et al. (2013) The dental plaque microbiome in health and disease. PLoS One 8: e58487.
- Edlund A, Yang Y, Yooseph S, Hall AP, Nguyen DD, et al. (2015) Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J 9: 2605-2619.
- Kuribayashi M, Kitasako Y, Matin K, Sadr A, Shida K, et al. (2012) Intraoral pH Measurement of Carious Lesions With qPCR of Cariogenic Bacteria to Differentiate Caries Activity. J Dent 40: 222-228.
- Yang F, Zeng X, Ning K, Liu KL, Lo CC, et al. (2012) Saliva microbiomes distinguish caries-active from healthy human populations. ISME J 6: 1-10.
- Takahashi N, Yamada T (2000) Glucose metabolism by Prevotella intermedia and Prevotella nigrescens. Oral Microbiol Immunol 15: 188-195.
- Kianoush N, Adler CJ, Nguyen KA, Browne GV, Simonian M, et al. (2014) Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS One 9: e92940.
- Jiang S, Gao X, Jin L, Lo EC (2016) Salivary Microbiome Diversity in Caries-Free and Caries-Affected Children. Int J Mol Sci 17: E1978.
- Xiao C, Ran S, Huang Z, Liang J (2016) Bacterial Diversity and Community Structure of Supragingival Plaques in Adults with Dental Health or Caries Revealed by 16S Pyrosequencing. Front Microbiol 22: 1145.
- Li Y, Ku CY, Xu J, Saxena D, Caufield PW (2005) Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res 84: 559-564.
- Dige I, Grønkjær L, Nyvad B (2014) Molecular studies of the structural ecology of natural occlusal caries. Caries Res 48: 451-460.
- Young DA, Featherstone JD (2013) Caries management by risk assessment. Community Dent Oral Epidemiol 41: e53-63.
- Ortiz S, Herrman E, Lyashenko C, Purcell A, Raslan K, et al. (2019) Sex-specific Differences in the Salivary Microbiome of Caries-Active Children. J Oral Microbiol 11: 1653124.
- Darout IA, Albandar JM, Skaug N, W Ali RW (2002) Salivary Microbiota Levels in Relation to Periodontal Status, Experience of Caries and Miswak Use in Sudanese Adults. J Clin Periodontol 29: 411-420.
- Lukacs JR (2011) Sex Differences in Dental Caries Experience: Clinical Evidence, Complex Etiology. Clin Oral Investig 15: 649-656.
- Thompson J, Pikis A (2012) Metabolism of sugars by genetically diverse species of oral Leptotrichia. Mol Oral Microbiol 27: 34-44.
- Costalonga M, Herzberg MC (2014) The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett 162: 22-38.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...