Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2641-1725
Research Article(ISSN: 2641-1725) 
Docking Study of New Ortho-Phenylenediamine Derivatives as COVID-19 Protease Inhibitors Volume 5 - Issue 2
Abdul M Gbaj1*, Nisreen H Meiqal1, Inass A Sadawe1, Salah M Bensaber1, Abdulathim A A Alshoushan2, Massaud Salem Maamar3 and Anton Hermann4
- 1Department of Medicinal Chemistry, Faculty of Pharmacy, University of Tripoli, Libya
- 2Food and Drug Control Centre (LFDA), Tripoli, Libya
- 3Department of Zoology, Faculty of Science, Tripoli University, Libya
- 4Department of Biosciences, University of Salzburg, Salzburg, Austria
Received: March 29, 2020; Published: June 16, 2020
*Corresponding author: Abdul M Gbaj, Associate Professor of Genetics and Biochemistry, Department of Medicinal Chemistry, Faculty of Pharmacy, University of Tripoli, LibyaK
DOI: 10.32474/LOJMS.2020.05.000207
Abstract
A series of new ortho-phenylenediamine derivatives were designed. The crystal structure of the main protease monomer was used as a target protein for molecular docking of ortho- phenylenediamine derivatives and a protein-ligand interaction analysis was performed using Auto Dock 4.2 software. Based on the docking score and after additional three-dimensional similarity analysis, NHM7 [(10,10’-((1E,1’E)-(1,2-phenylenebis(azanylylidene))bis(methanylylidene))bis(anthracen-9(8aH)-one)] had the highest binding energy. The calculated binding energy of ortho- phenylenediamine indicated effective binding of the proposed inhibitors to COVID-19 proteinase.
Keywords: Coronavirus; severe acute respiratory syndrome; covid-19 protease inhibitors; lopinavir; n3 inhibitor; molecular docking
Introduction
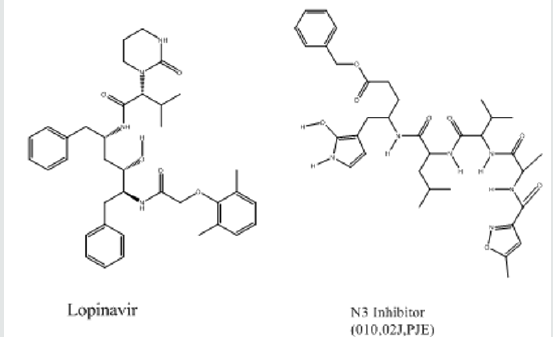
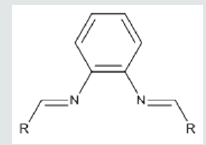
Coronaviruses (CoVs) comprise families of viruses pertained to the family of Coronaviridae. The viruses can circulate in humans and cause serious infections antinutrient the respiratory system [1,2]. Acute respiratory syndrome coronavirus (SARS-CoV) revealed that it can cause severe andoccasionallydeadly respiratory tract infections in humans [3-5]. The consecutive outbreaks, in addition, emphasize the threat of these viruses and caused a pandemic warning that has been declared a public health crisis of international anxiety [5-7].There are many potential targets against COVID-19 and among targets replication-related enzymes, such as protease3CL(pro) [also called SARS-CoV 3CL(pro)] are extremely conserved [8-10]. It has been reported that drugs that inhibit proteases are able preventing proliferation and replication of the virus by impeding with post-translational processing of vital viral polypeptides. In addition, they could also decrease the risk of drug-resistance produced by mutation. Following the protease inhibition approach two standard protease inhibitors were used as lead including Lopinavir and N3 (Figure 1) inhibitors identified of being capable to inhibit SARS-Covian protease [11]. It has been reported that the SARS-CoV main protease has 96.1% homology with the COVID-19main protease andthereforemay be utilized as a homologous target for screening of ortho- phenylenediamine derivatives that could inhibit the proliferation and replication of COVID-19 [12-15]. The ortho- phenylenediamine derivatives are Schiff bases recognized for their therapeutic value as they were reported to have anti-inflammatory, analgesic, antiviral, antitumor, antifungal and antibacterial properties [16-21]. Molecular modeling is a recognized computational toolto aid early drug discovery and development. It is used to generate ideasof a compounds or macromolecules 3D conformation, protein–ligand interactions, and allows forecasts about biological activities. The integration of molecular modeling in drug or vaccine design can help in early drug or vaccine discoveries [22-24]. The main aim of this study is to further identify protease as a target, and by computational drug repurposing procedures to allocate appropriate inhibitory agents.
Materials and Methods
Molecular docking
The starting geometry of the ortho-phenylenediamine derivatives was constructed using chem3D Ultra software (version 8.0, Cambridge soft Com., USA). The optimized geometry of orthophenylenediamine derivatives with the lowest energy was used for molecular dockings. The crystal structure of COVID-19 main protease in complex with the inhibitor N3 (6LU7) was downloaded from the Protein Data Bank http://www.rcsb.org/structure/6LU7 and COVID-19 and the main protease in apo form (6M03) was downloaded from the Protein Data Bank http://www.rcsb.org/ structure/6m03. Molecular dockings of ortho- phenylenediamine derivatives with 6LU7and 6M03 was accomplished by Auto Dock 4.2 software from the Scripps Research Institute (TSRI) ( http:// autodock.scripps.edu/). Firstly, polar hydrogen atoms were added into protein molecules. Then, partial atomic charges of the protease enzymes and ortho- phenylenediamine derivatives molecules were calculated using Kalman methods [25]. In the process of molecular docking, the grid maps of dimensions: (60Å X 60Å X 60Å) and (36.8Å X 64.6Å X 60Å) for 6LU7 and 6M03, respectively, with a gridpoint spacing of 0.376Å and the grid boxes is centered. The number of genetic algorithms runs and the number of evaluations were set to 100. All other parameters were default settings. Cluster analysis was performed on the basis of docking results by using a root mean square (RMS) tolerance of 2.0Å, dependent on the binding free energy. Lastly, the dominating configuration of the binding complex of ortho- phenylenediamine derivatives and protease enzymes fragments with minimum energy of binding were determined which relied strongly on the information of 3D-structures of the protease binding site and ultimately generated a series of proteasebinding complexes.
Results and Discussion
Molecular docking analysis
Table 1 shows the binding energies of Lopinavir, N3 (Figure 1, as standards), ortho-phenylenediamine derivatives, and protease enzymes (6LU7 and 6M03) obtained by the molecular docking strategy. Molecular dockings of the ortho-phenylenediamine derivatives with protease enzymes (6LU7 and 6M03) were performed using Auto Dock 4.2 to obtain information about interaction forces between ortho phenylenediamine derivatives and protease enzymes (6LU7 and 6M03). ortho-phenylenediamine derivatives and protease enzymes (6LU7 and 6M03) were kept as flexible molecules and were docked into seven forms of rigid protease enzymes (6LU7 and 6M03) to obtain the preferential binding site to ortho-phenylenediamine derivatives on protease enzymes (6LU7 and 6M03). The molecular docking results are shown in Table 1. The modeling studies indicate an der Waals, hydrogen bonding (Table 1) and electrostatic interactions between ortho-phenylenediamine derivatives with protease enzymes (6LU7 and 6M03). The contribution of van der Waals and hydrogen bonding interaction is much greater than that of the electrostatic interaction because the sum of van der Waals energy, hydrogen bonding energy and desolation free energy is larger than the electrostatic energy, [26,27].The ortho-phenylenediamine derivatives, and protease enzymes (6LU7 and 6M03) interactions are shown in Figure 2. Ortho phenylenediamine derivatives provide higher binding energy (-8.1 to -11.0kcal/mol) compared to standard 6LU7 and 6M03 (-7.0 to -7.9 kcal/mol) (Table 1 & Figure 2) indicates four hydrogen bonds between NHM7 and 6LU7.In addition, NHM7 showed good docking interaction of -11.0 kcal/mol with the 6LU7 binding site (GLU166, VAL3, GLU166and LEU4) (Figure 2). Compound NHM7 has the highest binding energy of the series. This compound has an extra phenyl moiety attached to the naphthyl analogue of the phenylenediamine Schiff`s base derivative with a log P value of 7.49 indicating the importance of the lipophilicity for the interaction with the active site. The interaction of similar Schiff’s base ortho phenylenediamine derivatives with the proteases binding site of the enzyme is essential for effective inhibition as previously reported [28-31]. Therefore, ortho-phenylenediamine derivatives may be considered the most effective NHM7 and 6LU7 proteases inhibitors. The obtained results using computational drug repurposing is an efficient way to find novel applications for already known drugs [32]. Molecular docking and binding free energy calculations for ortho- phenylenediamine derivatives can be used to forecast drugtarget interactions and binding affinity (Figure 3). The appearance of resistance to existing antiviral drugs or vaccines is a major challenge in antiviral drug development. The drug repurposing technique allows finding novel antiviral agents within a short period in order to overcome the challenges in antiviral therapy. Computational drug repurposing has previously been used to recognize drug candidates for viral infectious diseases like ZIKA, Ebola, influenza and dengue infections. These methods were also utilized to recognize possible drugs against MERS-CoV and SARSCoV [33,34] and following the COVID-19 outbreak, computational repurposing has been and are used for COVID-19.
Figure 2: Interaction model between NHM7 with COVID-19 main protease (6LU7) active site. NHM7 l green colour. Hydrogen bonds green broken line.
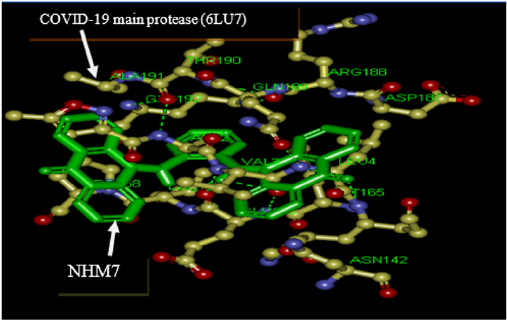
Table 1: Various energies in the binding process of ortho-phenylenediamine derivatives, N3 and Lopinavir with COVID-19 protease enzymes (6LU7, 6M03) obtained from molecular docking. The unit of all energies (ΔG) is kcal/mol.

Conclusion
In spite of the economic and societal shock of COVID-19 infections and the probability of future outbreaks of even more stern pathogenic COVID-19 in humans, there is still a lack of efficient antiviral strategies to treat COVID-19 and only few options are available to prevent COVID-19 infections. Rapid development and use of a broad-spectrum protease inhibitor alone or in combination with other potent inhibitors of proteases might fill the therapeutic gap spanning quarantine and hospital setting. Further elaborative work is necessary for better understanding the mechanisms of protease inhibition. According to modeling studies ortho-phenylenediamine derivatives may have the ability to inhibit COVID-19 proteases making them reasonable candidates for consideration of clinical trials and warrant further examination. Results presented in this study shall motivate future efforts in finding potent ortho-phenylenediamine derivatives that can be used for COVID-19 protease inhibition in vivo.
Acknowledgement
The authors gratefully acknowledge the technical support and valuable suggestions obtained from MS Amira Abdul Gbaj (Novelien Zone, Tripoli, Libya).
References
- Li LQ, HuangT, WangYQ, WangZP, LiangY, et al. (2020) novel coronavirus patients' clinical characteristics, discharge rate and fatality rate of meta-analysis. J Med Virol.
- Kim CJ (2020) New Year and coronavirus. J Exerc Rehabil 16(1):1.
- Chen Q, Quan B, Li X, G Gao,Zheng W (2020) A report of clinical diagnosis and treatment of 9 cases of coronavirus disease 2019. J Med Virol 92(6): 683-687.
- MaffioliEM (2020) How Is the World Responding to the 2019 Coronavirus Disease Compared with the 2014 West African Ebola Epidemic? The Importance of China as a Player in the Global Economy. Am J Trop Med Hyg 102(5):924-925.
- Li W, Cui H, Li K, Fang Y, Li S (2020) Chest computed tomography in children with COVID-19 respiratory infection. Pediatr Radiol 50(6):796-799.
- Zhao S, Cao P, Gao D, Zhuang Z, Cai Y, et al. (2020) Serial interval in determining the estimation of reproduction number of the novel coronavirus disease (COVID-19) during the early outbreak. J Travel Med.
- Qin C,Zhou L,Zh U,Zhang S, Yang S, et al. (2020) Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis.
- Holmes KV,Dominguez SR (2013) The new age of virus discovery: genomic analysis of a novel human betacoronavirus isolated from a fatal case of pneumonia. mBio 4(1):e00548-12.
- Chan JF,Lau SK, PC Woo (2013) The emerging novel Middle East respiratory syndrome coronavirus: the "knowns" and "unknowns". J Formos Med Assoc112(7):372-381.
- Zumla A, Azhar ER,Arabi Y, Alotaibi B,Rao M, et al. (2015) Host-directed therapies for improving poor treatment outcomes associated with the middle east respiratory syndrome coronavirus infections. Int J Infect Dis 40:71-74.
- Nukoolkarn V, Lee VS,Malaisree M,Aruksakulwong O, Hannongbua S (2008) Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J Theor Biol 254(4):861-867.
- Ton AT,Gentile F, Hsing M,Ban F, Cherkasov A (2020) Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol Inform.
- Kandeel M,Ibrahim AA, Fayez M,Al Nazawi M (2020) From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and non-structural genes. J Med Virol 92(6): 660-666.
- Morse JS,Lalonde T,Xu S,Liu WR (2020) Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. Chembiochem 21(5):730-738.
- Zhou J,Fang L,Yang Z,Xu S,Lv M (2019) Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB J33(12):14575-14587.
- MnguniMJ, A Lemmerer (2015)A structural study of 4-aminoantipyrine and six of its Schiff base derivatives. Acta Crystallogr C Struct Chem71(Pt 2):103-109.
- Anupama B,Sunita M,Shiva LD, Ushaiah B,Gyana KC (2014) Synthesis, spectral characterization DNA binding studies and antimicrobial activity of Co(II)Ni(II) Zn(II) Fe(III) and VO(IV) complexes with 4-aminoantipyrine Schiff base of ortho-vanillin. J Fluoresc 24(4):1067-1076.
- Raman N,Sakthivel A, Pravin N (2014) Exploring DNA binding and nucleolytic activity of few 4-aminoantipyrine based amino acid Schiff base complexes: a comparative approach. Spectrochim Acta A Mol Biomol Spectrosc125:404-413.
- Selwin JR,Shiju C,Joseph J,Justin DC, Arish D (2014) Synthesis and characterization of metal complexes of Schiff base ligand derived from imidazole2carboxaldehyde and 4aminoantipyrine. Spectrochim Acta A Mol Biomol Spectrosc133:149-155.
- GopalakrishnanS,JosephJ (2009) Antifungal Activities of Copper(II) with Biosensitive Macrocyclic Schiff Base Ligands Derived from 4-Aminoantipyrine Derivatives. Mycobiology37(2):141-146.
- Chandra S,Jain D,Sharma AK, P Sharma (2009) Coordination modes of a schiff base pentadentate derivative of 4-aminoantipyrine with cobalt(II), nickel(II) and copper(II) metal ions: synthesis spectroscopic and antimicrobial studies. Molecules 14(1):174-190.
- Chowdhury S, Happonen L, Khakzad H,Malmstrom L,Malmstrom J (2020) Structural proteomics, electron cryo-microscopy and structural modeling approaches in bacteria-human protein interactions. Med Microbiol Immunol 209(3): 265-275.
- John R, Arango-Jaramillo S, Self S,SchwartzDH (2004) Modeling partially effective HIV vaccines in vitro. J Infect Di189(4):616-623.
- Verlinde CL, Merritt EA, Van denAF, Kim H, Feil I, et al. (1994) Protein crystallography and infectious diseases. Protein Sci3(10):1670-1686.
- Tiwari R, Mahasenan K, Pavlovicz R, Li C,Tjarks W(2009) Carborane clusters in computational drug design: a comparative docking evaluation using AutoDock, FlexX, Glide, and Surflex. J Chem Inf Model 49(6):1581-1589.
- Holt PA,Chaires JB, TrentJO (2008) Molecular docking of intercalators and groove-binders to nucleic acids using Autodock and Surflex. J Chem Inf Model48(8):1602-1615.
- Gilad Y, Senderowitz H (2014) Docking studies on DNA intercalators. J Chem Inf Model 54(1):96-107.
- Konarikova K,Frivaldska J, Gbelcova H,Sveda M, Ruml T, et al (2019) Schiff base Cu(II) complexes as inhibitors of proteasome in human cancer cells. Bratisl Lek Listy120(9):646-649.
- Billinger E,Zuo S, Johansson G (2019) Characterization of Serine Protease Inhibitor from Solanum tuberosum Conjugated to Soluble Dextran and Particle Carriers. ACS Omega 4(19):18456-18464.
- Miranda PHA, Lacerda KCD, Araujo CM,Barichello JM,Lima WG (2018) Oral formulation of DPP-4 inhibitor plus Quercetin improves metabolic homeostasis in type 1 diabetic rats. Sci Rep 8(1):15310.
- Patil RH, Kalam Khan FA, Jadhav K, Damale M,Akber AS, et al. (2018) Fungal biofilm inhibition by piperazine-sulphonamide linked Schiff bases: Design, synthesis, and biological evaluation. Arch Pharm Weinheim351(3-4):e1700354.
- Vanhaelen Q,Mamoshina P, Aliper AM, Artemov A,Lezhnina K, et al. (2017) Design of efficient computational workflows for in silico drug repurposing. Drug Discov Today22(2):210-222.
- Dyall K,Coleman CM, Hart BJ, Venkataraman T,Holbrook MR, et al. (2014) Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother 58(8):4885-4893.
- Dyall J, Gross R,Kindrachuk J,Johnson RF, Olinge CG, et al. (2017) Middle East Respiratory Syndrome and Severe Acute Respiratory Syndrome: Current Therapeutic Options and Potential Targets for Novel Therapies. Drugs77(18):1935-1966.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...