Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4544
Research Article(ISSN: 2637-4544) 
UBE2C Promotes Breast Cancer Cell Proliferation through Upregulation of Bcl2 Volume 5 - Issue 1
Changjiang Yu1, Yangling Zhang2, Jingjing Huang2, Shengli Zeng1, Fan Chen1 and Jiangming Xiang2*
- 1Department of Breast Surgery, Chongqing Rongchang Health Center for Women and Children, China
- 2Department of Breast Surgery, Attending Physician of Breast Surgery in Chongqing Changshou Maternal and Child Health Care Hospital, China
Received:January 14, 2022; Published: January 21, 2022
Corresponding author: Jiangming Xiang, Department of Breast Surgery, Attending Physician of Breast Surgery in Chongqing Changshou Maternal and Child Health Care Hospital, China
DOI: 10.32474/IGWHC.2022.05.000205
Abstract
Purpose: To investigate the mechanism by which UBE2C affects breast cancer tumor progression through regulation of Bcl2.
Methods: qRT-PCR to detect the expression of UBE2C in breast cancer cells MCF-7 and normal breast epithelial cells MCF-10A; Western blot to detect the expression of UBE2C and Bcl2 protein in breast cancer cells; CCK8 cell viability assay and clone formation assay by plasmid or siRNA transfection to overexpress or knockdown UBE2C proliferation of breast cancer cells; Co-IP to verify the interaction between UBE2C and Bcl2.
Results: UBE2C and Bcl2 were significantly highly expressed in breast cancer cells; overexpression of UBE2C promoted breast cancer cell proliferation and knockdown of UBE2C inhibited breast cancer cell proliferation; UBE2C interacted with Bcl2, and UBE2C promoted breast cancer cell proliferation by promoting Bcl2 expression and thus breast cancer cell proliferation.
Conclusion: UBE2C promotes breast cancer cell proliferation through upregulation of Bcl2.
Keywords: Breast Cancer; UBE2C; Bcl2; Proliferation
Introduction
Breast cancer is the most prevalent malignant tumor in women, and it is also the cause of the second highest death rate among cancer patients. Tumor-associated macrophages (TAMs) are the most common immune cells in the breast tumor microenvironment, and they govern breast cancer progression [1]. In the year 2020, 685,000 people will have died from BC, out of a total of 2.3 million cases identified worldwide. A total of 7.8 million women were diagnosed with BC in the previous 5 years by the end of 2020 [2]. Out of a total of 2.3 million cases diagnosed worldwide, 685,000 people will have died from BC by 2020. By the end of 2020, 7.8 million women will have been diagnosed with BC in the previous five years [3]. Breast cancer has been classified as luminal A, luminal B, HER2-positive, and basal-like triple negative breast cancer (BL/TNBC) using histological categorization based on the expression of Estrogen Receptor (ER), Progesterone Receptor (PR), and/or human epidermal growth factor receptor-2 (HER2) [4]. Luminal cancer has the largest incidence (70%) and is followed by HER2-positive (15-20%) and triple-negative (15%) tumors, with triple-negative malignancies having the highest recurrence rate. Endocrinology-based therapy and chemotherapy are utilized to treat luminal tumors, depending on the tumor’s response [5].
TNBC is mostly treated with chemotherapy, with just a few alternatives for PARP inhibitors or immunotherapy [6]. Almost of refractory breast tumors develop resistance to second-line treatments at some point [7]. Chemotherapy is an important part of cancer treatment that can cause nausea, peripheral neuropathy, and a variety of organ damage. However, the most serious side effect of chemotherapy is cognitive impairment, often known as chemo brain or CICI (Chemotherapy-Induced Cognitive Impairment) [8]. Women with breast cancer need personal health-related tools because they assist them preserve their own health and well-being during various stages of their illness [9]. In addition, the discovery of the molecular basis of breast cancer has led to the development of gene therapy as a viable treatment option for this disease. Gene therapy is inserting genetic material into target cells via a vector, followed by gene correction, addition, or suppression. It is crucial to target tumor cells while avoiding normal cells in this procedure [10]. Randomized Control Trials (RCTs), observational research, and computer model data all support mammography screening. Digital breast tomosynthesis is a new technology that improves the sensitivity and specificity of mammography by addressing constraints caused by overlapping breast tissue. [11]. Currently, the number of Circulating Tumor Cell (CTC) is a prognostic indicator of breast cancer overall survival, and Tumor Mutation Burden (TMB) can be used checkpoint inhibitors. Currently, clinical methods such as Polymerase Chain Reaction (PCR) and Next Generation Sequencing (NGS) are mainly adopted to evaluate these biomarkers, which are time-consuming and expansive [12]. Growing evidence has demonstrated that UBE2C plays a critical role in cancer progression, but there is no study focusing on the prognosis, upstream regulation mechanism, and immunological roles of UBE2C across diverse tumor types [13]. In this study, we found that UBE2C was elevated in this human pan-cancer analysis and that high expression of UBE2C was associated with poor prognosis. Finally, we showed that downregulation of UBE2C decreased cell viability and slowed proliferation of breast cancer cells. Our study elucidates that UBE2C promotes breast cancer cell proliferation through regulation of Bcl2, and that overexpression of Bcl2 reverses the inhibitory effect of downregulation of UBE2C on breast cancer cell proliferation and provides a prognostic indicator as well as a promising therapeutic target for breast cancer patients.
Experimental Method
Cell culture and reagents
Human normal breast epithelial cells MCF-10A and breast cancer cells MCF-7 were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured with RPMI-1640 medium (Hy Clone, Logan City, UT) containing 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 μg/ml streptomycin (Gibco, Carlsbad, CA, USA), and the culture flasks were placed in a constant temperature incubator at 37°C containing 5% CO2 [14,15].
Plasmid, siRNA transfection
Plasmids or siRNAs were transfected in breast cancer cells in the following groupings: oe-NC, oe-UBE2C, si-NC, si-UBE2C, si-UBE2C + oe-Bcl2. 1 × 105 cells were inoculated in each well of a six-well plate, and when cell confluency reached 60-70%, 750μl optimem culture was first added to each well, and 125μl optimem and 5μl Lipo3000 (L3000001 , Invitrogen, Carlsbad, CA, USA) mixed and left for 5 min, then 125μl optimem and plasmid/siRNA mixed and left for 5 min, then both were mixed and left for 20 min, 250μl of the solution was added to the wells, slightly shaken well, placed in the incubator, and the solution was changed after 16 h. After 48hr, cells could be extracted RNA, and cellular proteins can be extracted after 72 h for subsequent experiments [16].
qRT-PCR
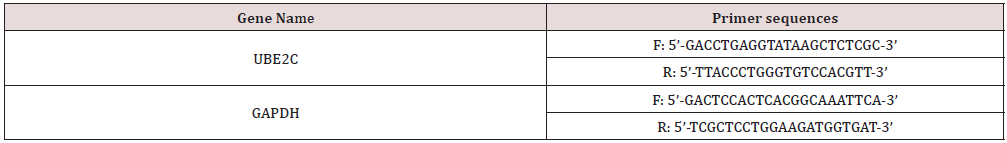
Total intracellular RNA was extracted using Trizol reagent (15596026, Invitrogen, USA) according to the manufacturer’s instructions, followed by reverse transcription of RNA to cDNA following the procedure of the Prime Script RT reagent Kit (RR047A, Takara, Japan) kit, and finally using the Fast SYBR Green PCR kit (Applied biosystems) with ABI PRISM 7300 RT-PCR system (Applied biosystems) was used to quantify the RNA, and three secondary wells were repeated for each sample. GAPDH was used as an internal reference. The relative gene expression was analyzed by the 2 -ΔΔCt method, calculated as △Ct = CT(target gene)-CT(Internal reference), △△Ct = △Ct(test group)-△Ct(control group), and the average value was taken after three replicate experiments. All primers were purchased from Shanghai Biotech Biology, and the primer sequences were as follows Table 1.
Western blot
The cells were removed, the cell surface was washed twice with PBS, 60μl of RIPA lysate + 1% protease inhibitor + 1% phosphatase inhibitor (Beyountian Biotechnology, Shanghai, China) was added and the cells were scraped off and lysed on ice for 45 min. after centrifugation at 4°C at 12000×g for 15 min, the protein supernatant was aspirated and the protein concentration was measured using a BCA kit (Beyountian Biotechnology, Shanghai, China). , Shanghai, China) to detect the protein concentration. Add 1/4 volume of 5×Loading Buffer (Beyountian Biotechnology, Shanghai, China) to the protein solution and boil for 10 min at 100°C to denature the proteins. 20 μg of each protein solution was separated by electrophoresis on a 10% SDS-PAGE gel. The strips were incubated overnight at 4℃ (12-16h) and washed three times with TBST solution for 5min each time after incubation. The strips were washed three times again with TBST solution after incubation for 1hr. ECL working solution (BM101, Biomiga, USA) was added dropwise to the strips and developed after 1min using a Bio Spectrum 600 imaging system (Ultra-Violet Products, UK). Protein strip gray values were calculated using Image J software, and the target protein gray value/GAPDH gray value was used as the relative protein expression. The experiments were repeated three times.
CCK8 cell viability assay
After digestion, the cells were centrifuged and the cell suspension was added to 96-well plates with 5×103cells per well, and 5 replicate wells were set up for each group. 24h later, the old medium was discarded and trastuzumab solution was added to each group for 0, 12, 24, 36, 48 and 60h. 10μl CCK8 reagent was added at the corresponding time points, and the absorbance OD value was detected at 450 nm using an enzyme marker. Cell viability was calculated by Graph Pad Prism7 software. The experiment was repeated three times [17,18].
Clone formation experiment
After transfection, the cells were seeded into six-well plates with 5×102 cells per well and incubated for two weeks. Cells were then fixed in methanol and then stained with crystal violet (Sigma- Aldrich). The number of colonies containing more than 50 cells was counted manually [19].
Immunoprecipitation (Co-IP)
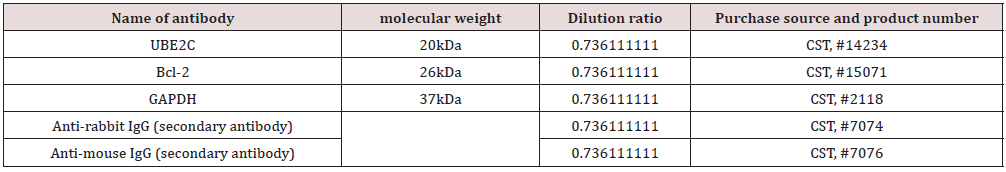
Cells were lysed with IP lysis buffer (Thermo Fisher, Inc) containing protease inhibitor (Sigma) and PMSF (Sigma) at 4°C for 1h. After centrifugation at high speed for 20 min, the supernatant was incubated with primary antibody and protein magnetic bead A (Invitrogen) for 4h at 4°C in a rotating mix, and then the beads were washed four times with IP lysis buffer. The beads were then eluted with 1×SDS, and the eluate was quantified by protein BCA, and the immunoprecipitation results of the two proteins were detected by Western blot. Primary antibody: UBE2C (20 kDa, 1:30, ab252940, Abcam, USA), Bcl2 (26 kDa, #15071, Cell Signaling Technology, MA, USA) [20].
Statistical analysis
Statistical analyses were performed using SPSS 21.0 software (IBM, Armonk, NY, USA). Measured data were expressed as mean ± standard deviation, and tests for normality and chi-squareness were performed. Paired t-tests were used to compare data within groups with normal distributions and equal variances, and unpaired t-tests were used to compare data between two groups. Data were compared between multiple groups by one-way Analysis of Variance (ANOVA) and Tukey’s post hoc test. Data from two groups at different time points were compared by repeated measures ANOVA. When data did not show normal distribution or equal variance, rank sum tests were performed. p < 0.05 was considered statistically significant.
Prediction of Results in the Literature
UBE2C is highly expressed in breast cancer cells
The expression levels of UBE2C were examined in human normal breast epithelial cells MCF-10A and breast cancer cells MCF-7. qRT-PCR and Western blot showed that the expression of UBE2C was significantly higher in MCF-7 cells than in MCF-10A cells (Figures 1A-B).
Figure 1: UBE2C is highly expressed in breast cancer cells. 1A: qRT-PCR assay of mRNA levels of UBE2C in MCF-10A and MCF-7 cells; 1B: Western blot assay of protein levels of UBE2C in MCF-10A and MCF-7 cells; Cell experiments were repeated three times.

Overexpression of UBE2C promotes proliferation of breast cancer cells
To understand the regulatory role of UBE2C in breast cancer, we transfected overexpressed or knocked down UBE2C by plasmid or siRNA. qRT-PCR and Western blot detected the transfection efficiency, and the results showed that: compared with the oe- NC group, the expression of UBE2C increased in the oe-UBE2C group; compared with the si-NC group, the expression of UBE2C in the si-UBE2C group The results of CCK8 cell viability assay and clone formation assay showed that the cell viability increased and the proliferation rate accelerated after overexpression of UBE2C, while the cell viability decreased and the proliferation rate slowed down after knockdown of UBE2C (Figures 2A-D). The above results indicated that overexpression of UBE2C could promote the proliferation of breast cancer cells, and downregulation of UBE2C could lead to the opposite result.
Figure 2: 2A: qRT-PCR to detect the transfection efficiency of overexpressing or knocking down UBE2C in MCF-7 cells; 2B: CCK8 cell viability assay to detect the cell viability of each group of cells at 0h, 12h, 24h, 36h and 48h; 2C: clone formation assay to detect the number of cell colonies formed in each group of cells after 48h of culture; The cell experiments were repeated three times. Figures 2: Overexpression of UBE2C promotes proliferation of breast cancer cells.

UBE2C promotes Bcl2 protein expression
To further investigate the role of UBE2C, we analyzed differentially expressed genes in MCF-7 cells with knockdown of UBE2C using microarray, and we screened a total of 269 differentially expressed genes (Figure 3A) by setting a screening threshold of Log FC > 2.0 and adj P value < 0.05, and the heatmap demonstrates the top 30 differentially expressed genes (Figure 3B), among which the expression of Bcl-2 was most significantly reduced. To determine whether UBE2C interacted with Bcl2 in breast cancer cells, we performed Co-IP assays in MCF-7 cells, and the results showed that there was specific binding between UBE2C and Bcl2 (Figure 3A). Western blot assay showed that the expression of Bcl2 was significantly higher in MCF-7 cells than in MCF-10A cells (Figure 3B). And Bcl2 protein expression increased after overexpression of UBE2C; Bcl2 protein expression decreased after knockdown of UBE2C (Figure 3C). The above results indicated that UBE2C could promote Bcl2 protein expression.
Figure 3: Figure Note: A: microarray analysis of differentially expressed genes in MCF-7 cells with knockdown of UBE2C, volcano plot showing differentially expressed genes; B: heat map showing the top 30 differentially expressed genes; C: Co-IP assay to verify that UBE2C interacts with Bcl2; D: Western blot detection of Bcl2 in MCF-10A and MCF-7 cells; E: Western blot to detect the protein level of Bcl2 after overexpression or knockdown of UBE2C. Cell experiments were repeated three times. Figures 2: UBE2C promotes Bcl2 protein expression.

UBE2C promotes breast cancer cell proliferation through upregulation of Bcl2
We then further explored whether UBE2C affects breast cancer cell proliferation by regulating Bcl2. After downregulating UBE2C in MCF-7 cells and then overexpressing Bcl2, Western blot assay results showed that UBE2C and Bcl2 protein levels were decreased in the si-UBE2C group compared with the si-NC group; compared with the si-UBE2C group, UBE2C protein levels were decreased in the si-UBE2C+oe-Bcl2 group and Bcl-2 protein CCK8 cell viability assay and clone formation assay showed that the cell viability was reduced and the proliferation rate was slowed down after downregulation of UBE2C; overexpression of Bcl2 on top of this group restored cell viability and cell proliferation rate (Figures 4A-4C). The above results indicated that UBE2C promoted the proliferation of breast cancer cells by regulating Bcl2, and overexpression of Bcl2 could reverse the inhibitory effect of downregulation of UBE2C on the proliferation of breast cancer cells.
Figure 4: Figure Note: 4A: Western blot to detect the protein levels of UBE2C and Bcl2 in each group of cells; 4B: CCK8 cell viability assay to detect the cell viability of each group of cells at 0h, 12h, 24h, 36h and 48h; 4C: clone formation assay to detect the number of cell colonies formed after 48h of culture in each group of cells. The cell experiments were repeated three times. Figures 4: UBE2C promotes breast cancer cell proliferation through upregulation of Bcl2.

Discussion
Breast cancer is considered one of the utmost neoplastic diseases globally, with a high death rate of patients. The incidence of BC in 2020 is the highest while the mortality rate is only fifth in the world, highlighting the need to prevent and treat BC [21]. Over the last decades, many approaches have been studied to early diagnose and treat it, such as chemotherapy, hormone therapy, immunotherapy, and MRI and biomarker tests; do not show the optimal efficacy [22]. These existing treatments for breast cancer are accompanied by serious side effects, so recognizing these challenges, medical researchers have made great efforts to find new ways to treat breast cancer, aiming to improve the quality of life and address the suffering of patients with breast cancer. Many diseases are accompanied by changes in certain biochemical indicators called biomarkers in cells or tissues. A variety of biomarkers, including proteins, nucleic acids, antibodies, and peptides, have been identified. Tumor biomarkers have been widely used in cancer risk assessment, early screening, diagnosis, prognosis, treatment, and progression monitoring [12]. Despite significant improvements in the way breast cancer is managed and treated, it continues to persist as a leading cause of death worldwide. If detected and diagnosed early, when tumors are small and localized, there is a considerably higher chance of survival. [23]. However, current detection and diagnostic methods lack the sensitivity and specificity necessary to identify breast cancer in the asymptomatic or very early stages. Therefore, there is a need to develop more rapid and reliable methods that can detect the disease earlier to improve disease management and patient outcomes.
Ubiquitin-conjugating enzyme E2C (UBE2C) has been shown to be associated with the occurrence of various cancers and involved in many tumorigenic processes. UBE2C is necessary for the correlated activity between mitotic cyclins and its substrates [24]. During the cell metaphase, UBE2C promotes cell cycle progression by interfering with cyclin B [25]. Our pan-cancer analysis indicated that UBE2C mRNA was decreased in the majority of normal tissues, while UBE2C was significantly elevated in cancer tissues, such as in colon cancer and lung cancer [26]. The high expression of UBE2C was associated with a poor prognosis in breast, colon, liver, lung, and ovarian cancer [27]. The aim of this study was to investigate the specific molecular mechanisms by which UBE2C affects the proliferation of Breast Cancer (BC) [28]. UBE2C has been described as a molecular marker for human lung cancer and liver cancer prognosis [29,30], while another study indicated that UBE2C could be a potential biomarker for tumorigenesis and prognosis in squamous cell carcinoma of the tongue [31]. In our study, we operated in a 4-step experiment. In the first step, the expression level of UBE2C was detected in human normal breast epithelial cells MCF-10A and breast cancer cells MCF-7. qRT-PCR and Western blot showed that the first conclusion that UBE2C was highly expressed in breast cancer cells was obtained. In the second step, to understand the regulatory role of UBE2C in breast cancer, we detected the expression level of UBE2C by plasmid or siRNA transfection to overexpress or knockdown UBE2C, and qRT-PCR and Western blot to detect the transfection efficiency, which confirmed that overexpression of UBE2C could promote the proliferation of breast cancer cells.
Next, to further investigate the role of UBE2C, we analyzed the genes differentially expressed in MCF-7 cells with knockdown UBE2C using microarray to determine whether UBE2C interacts with Bcl2 in breast cancer cells, we performed Co-IP assays in MCF- 7 cells, and the above results indicated that UBE2C promotes Bcl2 protein expression. Finally, we further explored whether UBE2C affects breast cancer cell proliferation by regulating Bcl2, and the results of the exploration indicated that UBE2C promotes breast cancer cell proliferation by regulating Bcl2, and that overexpression of Bcl2 reverses the inhibitory effect of downregulating UBE2C on breast cancer cell proliferation. In a word, UBE2C mRNA and protein level were highly expressed in ESCC and UBE2C was likely to play different roles in different stages of the ESCC [32]. The above findings suggest that UBE2C plays an important role in cancer development and may be a promising pan-cancer biomarker for prognosis and treatment. UBE2C promotes breast cancer cell proliferation through upregulation of Bcl2, and UBE2C may serve as a promising target for developing therapeutic strategies for breast cancer.
Limitation
This study has several limitations. First, it lacks systematic quantitative analysis and there is some selection bias in the choice of cited literature in this review, which may lead to bias in the findings and conclusions of the study. It remains to be confirmed whether the findings are applicable to the entire global population. In the cytologic evaluation of small cell breast tumors, attention should be paid to the broad diversity of tumors and consideration of clinical, hematologic, and radiologic features. Finally, the prognostic and immunological role of UBE2C in human cancers has been an unknown area. Further understanding of the factors that stimulate tumor angiogenesis and the interactions between these new pathways and more established mediators may also allow for improved therapeutic approaches without the need for new drugs.
Conclusion
In summary, our study shows that UBE2C is elevated in human cancers and positively correlates with unfavorable prognosis. Furthermore, high expression of UBE2C was associated with plasmid or siRNA transfection, and Bcl2 protein expression degree. Finally, we identified the upstream regulatory mechanism of UBE2C in Bcl2, i.e., UBE2C promotes breast cancer cell proliferation by regulating Bcl2, and overexpression of Bcl2 reverses the inhibitory effect of downregulating UBE2C on breast cancer cell proliferation. Here, we provide the first evidence that UBE2C plays a key role in cancer progression and immune response in human pan-cancer through upregulation of Bcl2.
References
- Huang X, Cao J, Zu X (2021) Tumor-associated macrophages: An important player in breast cancer progression. Thoracic cancer.
- Rosenqvist H (1951) Breast cancer. Nordisk medicine 46: 1802.
- Sheikh A Md S, Kesharwani P J (2021) Aptamer grafted nanoparticle as targeted therapeutic tool for the treatment of breast cancer. Biomed Pharmacother 146: 112530.
- Ma R (2020) Monocyte-derived macrophages promote breast cancer bone metastasis outgrowth. J Exp Med 217(11): e20191820.
- Harbeck N, Gnant M (2017) Breast cancer. Lancet, London, England, UK 389(10074): 1134-1150.
- Cortesi L, Rugo H, Jackisch CJTo (2021) An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Target Oncol 16(3): 255-282.
- Warburg O, Wind F, Negelein E (1927) THE METABOLISM OF TUMORS IN THE BODY. The Journal of general physiology 8(6): 519-530.
- Sahu K, Langeh U, Singh C, Singh A (2021) Crosstalk between anticancer drugs and mitochondrial functions. Current research in pharmacology and drug discovery 2: 100047.
- Skyttä M, Pietilä A M, Stolt M, Kangasniemi M (2021) Identifying personal health-related resources of women with breast cancer for nursing: An integrative review. Scandinavian journal of caring sciences.
- Dastjerd N (2021) Gene therapy: A promising approach for breast cancer treatment.
- Rahman W T, Helvie M A (2021) Breast cancer screening in average and high-risk women. Best practice & research. Clinical obstetrics & Gynaecology.
- Xie X (2021) Evaluating Cancer-Related Biomarkers Based on Pathological Images: A Systematic Review. Front Oncol 11: 763527.
- Jiang X (2021) Comprehensive Pan-Cancer Analysis of the Prognostic and Immunological Roles of the METTL3/lncRNA-SNHG1/miRNA-140-3p/UBE2C Axis. Front Cell Dev Biol 9: 765772.
- Zeng J (2020) miR-204/COX5A axis contributes to invasion and chemotherapy resistance in estrogen receptor-positive breast cancers. Cancer Lett 492: 185-196.
- Pan X (2021) METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med 53: 91-102.
- Han M (2020) Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer 19: 26.
- Dong Huaying, Hu Jianguo, Zou Kejian, Ye Mulin, Chen Yuanwen, et al. (2019) Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol Cancer 18(1): 3.
- Tao L, Wu Y, Zhang S J N (2019) MiR-21-5p enhances the progression and paclitaxel resistance in drug-resistant breast cancer cell lines by targeting PDCD4. Neoplasma 66: 746-755.
- Zhao L (2019) LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. 10: 731.
- Song Z (2019) UCHL3 promotes pancreatic cancer progression and chemo-resistance through FOXM1 stabilization. Am J Cancer Res 9(9): 1970-1981.
- Shen L, Zhang S, Wang K, Wang X (2021) Familial Breast Cancer: Disease Related Gene Mutations and Screening Strategies for Chinese Population. Front Oncol 11: 740227.
- Madamsetty V (2021) Chitosan: A versatile bio-platform for breast cancer theranostics. Journal of Controlled Release.
- Hanna, K (2021) Raman spectroscopy: current applications in breast cancer diagnosis, challenges and future prospects. Br J Cancer 1-15.
- Aristarkhov A (1996) E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. 93: 4294-4299.
- Binné, U (2007) Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol 9: 225-232.
- Hao Z, Zhang H, Cowell J Medicine Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumour Biol 33(3): 723-730.
- van Ree J, Jeganathan K, Malureanu L, van Deursen JJTJocb (2010) Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. 188: 83-100.
- Lu Z, Song J, Sun T, Sun GJC (2021) UBE2C affects breast cancer proliferation through the AKT/mTOR signaling pathway. Chin Med J (Engl) 134(20): 2465-2474.
- Kadara H (2009) Identification of gene signatures and molecular markers for human lung cancer prognosis using an in vitro lung carcinogenesis system. 2: 702-711.
- Wu Y (2019) UBE2C Induces Cisplatin Resistance via ZEB1/2-Dependent Upregulation of ABCG2 and ERCC1 in NSCLC Cells. 8607859.
- Liu P (2020) UBE2C is a Potential Biomarker for Tumorigenesis and Prognosis in Tongue Squamous Cell Carcinoma. Diagnostics (Basel) 10(9): 674.
- Li R (2021) Overexpression of UBE2C in esophageal squamous cell carcinoma tissues and molecular analysis. BMC Cancer 21: 996.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...