Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-6062
Short communication(ISSN: 2638-6062) 
How the Artificial Intelligence Tool iPGK-PseAAC is Working in Predicting lysine Phosphoglycerylation Sites in Proteins Volume 4 - Issue 2
Kuo-Chen Chou*
- Gordon Life Science Institute, Boston, United States of America
Received: March 10, 2020; Published: March 17, 2020
*Corresponding author: Kuo-Chen Chou Gordon Life Science Institute, Boston, Massachusetts 02478, United States of America
DOI: 10.32474/PRJFGS.2020.04.000181
Short communication
In 2016 a very powerful AI (artificial intelligence) tool has been established for predicting lysine succinylation sites in proteins, one of the most important post modifications in proteins [1].
To see how the web-server is working, please do the following.
Step 1. Opening the web-server at you will see the top page of iSuc-PseOpt on your computer screen, as shown in Figure 1. Click on the Read Me button to see a brief introduction about this predictor.
Figure 1: A semi-screenshot for the top-page of the iSuc-PseOpt web-server at (Adapted from [1] with permission).
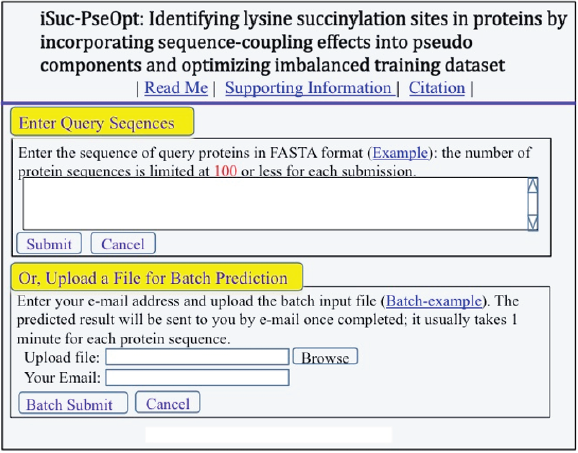
Step 2. Either type or copy/paste your query protein sequences into the input box at the center of Figure1. The input sequences should be in the FASTA format. For the examples of sequences in FASTA format, click the Example button right above the input box.
Step 3. Click on the Submit button to see the predicted result. For example, if you use the Sequences in the Example window as the input, after a few seconds, you will see the corresponding predicted results, which is fully consistent with experiment observations.
Step 4. Click the Data button to download the benchmark dataset used in this study.
Step 5. Click the Citation button to find the relevant papers that document the detailed development and algorithm for iSuc-PseOpt.
It is anticipated that the Web-Server will be very useful because the vast majority of biological scientists can easily get their desired results without the need to go through the complicated equations in [1] that were presented just for the integrity in developing the predictor. Also, note that the web-server predictor has been developed by strictly observing the guidelines of “Chou’s 5-steps rule” and hence have the following notable merits [2] and three comprehensive review papers [3-5] (1) crystal clear in logic development, (2) completely transparent in operation, (3) easily to repeat the reported results by other investigators, (4) with high potential in stimulating other sequence-analyzing methods, and (5) very convenient to be used by the majority of experimental scientists. It has not escaped our notice that during the development of iDNA6mA-PseKNC web-server, the approach of general pseudo amino acid components [6] or PseAAC [7] had been utilized and hence its accuracy would be much higher than its counterparts, as concurred by many investigators [8, 9]. For the marvelous and awesome roles of the “5-steps rule” in driving proteome, genome analyses and drug development, see a series of recent papers [10-31]where the rule and its wide applications have been very impressively presented from various aspects or at different angles.
References
- Jia J, Liu Z, Xiao X, B Liu, KC Chou (2016) iSuc-PseOpt: Identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal Biochem 497: 48-56.
- Barukab O, Khan YD, Khan SA, Chou KC (2019) iSulfoTyr-PseAAC: Identify tyrosine sulfation sites by incorporating statistical moments via Chou's 5-steps rule and pseudo components Current Genomics. Curr Genomics 20(4): 306-320.
- Chou KC (2011) Some remarks on protein attribute prediction and pseudo amino acid composition (50th Anniversary Year Review, 5-steps rule). J Theor Biol 273: 236-247.
- Chou KC (2019) Advance in predicting subcellular localization of multi-label proteins and its implication for developing multi-target drugs. Curr Med Chem 26: 4918-4943.
- Chou KC (2019) Impacts of pseudo amino acid components and 5-steps rule to proteomics and proteome analysis.Curr Top Med Chem 19(25): 2283-2300.
- Chou KC (2001) Prediction of protein cellular attributes using pseudo amino acid composition. Proteins 43(3): 246-255.
- Chou KC (2005) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21(1): 10-19.
- Kabir M, Ahmad S, Iqbal M, Hayat M (2020) iNR-2L: A two-level sequence-based predictor developed via Chou's 5-steps rule and general PseAAC for identifying nuclear receptors and their families. Genomics 112(1): 276-285.
- Malebary SJ, Rehman MSU, Khan YD (2019) iCrotoK-PseAAC: Identify lysine crotonylation sites by blending position relative statistical features according to the Chou's 5-step rule. PLoS One 14(11): e0223993.
- Chou KC (2019) The cradle of Gordon Life Science Institute and its development and driving force. Int J Biol Genetics 1: 1-28.
- Chou KC (2019) Showcase to illustrate how the web-server iDNA6mA-PseKNC is working. Journal of Pathology Research Reviews & Reports 1: 1-15.
- Chou KC (2019) The pLoc_bal-mPlant is a Powerful Artificial Intelligence Tool for Predicting the Subcellular Localization of Plant Proteins Purely based on their Sequence Information. Int J Nutr Sci 4(1): 1-4.
- Chou KC, Cheng X, Xiao X (2019) pLoc_bal-mEuk: predict subcellular localization of eukaryotic proteins by general PseAAC and quasi-balancing training dataset. Med Chem 15(5): 472-485.
- Chou KC (2019) Showcase to illustrate how the web-server iNitro-Tyr is working. Glo J of Com Sci and Infor Tec 2: 1-16.
- Chou KC (2019) Gordon Life Science Institute: Its philosophy, achievements, and perspective. Annals of Cancer Therapy and Pharmacology 2: 1-26.
- Chou KC (2020) Showcase to Illustrate how the webserver pLoc_bal-mEuk Is working. Biomed J Sci & Tech Res 24.
- Chou KC (2020) The pLoc_bal-mGneg Predictor is a Powerful Web-Server for Identifying the Subcellular Localization of Gram-Negative Bacterial Proteins based on their Sequences Information Alone. ijSci 9: 27-34.
- Chou KC (2020) How the artificial intelligence tool iRNA-2methyl is working for RNA 2’-Omethylation sites. Journal of Medical Care Research and Review 3: 348-366.
- Chou KC (2020) Showcase to illustrate how the web-server iKcr-PseEns is working. Journal of Medical Care Research and Review 3: 331-347.
- Chou KC (2020) The pLoc_bal-mVirus is a powerful artificial intelligence tool for predicting the subcellular localization of virus proteins according to their sequence information alone. J Gent & Genome 4.
- Chou KC (2019) How the artificial intelligence tool iSNO-PseAAC is working in predicting the cysteine S-nitrosylation sites in proteins. J Stem Cell Res Med 4: 1-9.
- Chou KC (2020) Showcase to illustrate how the web-server iRNA-Methyl is working. J Mol Genet 3: 1-7.
- Chou KC (2020) How the Artificial Intelligence Tool iRNA-PseU is Working in Predicting the RNA Pseudouridine Sites. Biomed J Sci & Tech Res pp.
- Chou KC (2020) Showcase to illustrate how the web-server iSNO-AAPair is working. J Gent & Genome 4.
- Chou KC (2020) The pLoc_bal-mHum is a Powerful Web-Serve for Predicting the Subcellular Localization of Human Proteins Purely Based on Their Sequence Information. Adv Bioeng Biomed Sci Res 3: 1-5.
- Chou KC (2020) Showcase to Illustrate How the Web-server iPTM-mLys is working. Infotext Journal of Infectious Diseases and Therapy [IJID] 1: 1-16.
- Chou KC (2020) The pLoc_bal-mGpos is a powerful artificial intelligence tool for predicting the subcellular localization of Gram-positive bacterial proteins according to their sequence information alone. Glo J of Com Sci and Infor Tec 2: 01-13.
- Chou KC (2020) Showcase to illustrate how the web-server iPreny-PseAAC is working. Glo J ofCom Sci and Infor Tec 2: 1-15.
- Chou KC (2020) Some illuminating remarks on molecular genetics and genomics as well as drug development. Molecular Genetics and Genomics 295: 261-274.
- Chou KC (2020) The Problem of Elsevier Series Journals Online Submission by Using Artificial Intelligence. Natural Science 12(2): 37-38.
- Chou KC (2020) The Most Important Ethical Concerns in Science. Natural Science 12: 35-36.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...




