Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2643-6744
Research ArticleOpen Access
Design of an Economic Model for Protectively Sharing Biomedical Data Volume 1 - Issue 4
Adebayo OT1* and Fasidi FO2
- 1Department of Information Technology, The Federal University of Technology Akure, Nigeria
- 2Department of Computer Science, The Federal University of Technology Akure, Nigeria
Received:August 13, 2019; Published: August 27, 2019
*Corresponding author: Adebayo OT, Department of Information Technology, The Federal University of Technology Akure, Nigeria.
DOI: 10.32474/CTCSA.2019.01.000117
Abstract
Sharing medical data (such as genomic) can lead to important discoveries in healthcare, but researches have shown that links between de-identified data and named persons are sometimes reestablished by users with malicious intents. Traditional approaches to curb this menace rely data use agreements, suppression and noise adding to protect the privacy of individual in the dataset, but this reduces utility of the data. Therefore, this paper proposed an economic game theoretic model design for quantifiable protections of genomic data. The model can be developed to find solution for sharing summary statistics under an economically motivated recipient’s (adversary) inference attack. The framework incorporates four main participants: Data Owners, Certified Institution (CI), Sharer and Researchers (Recipients). The data Sharer and Researcher (who are the players) are economically motivated.
Keywords: Game; Publisher; Recipient
Introduction
Using the massive amount of information encoded in the
biomedical can have significant effects on personalized medicine,
paternity testing and disease susceptibility testing. With data
analyses, vital information about an individual can be revealed,
for instance, disease susceptibility testing can determine if an
individual is likely to have a specific disease such as breast cancer
and diabetes or not [1]. In personalized medicine, a physician
can prescribe a safe and effective medical treatment built on the
patient’s genetic profile to minimize side effects. The increasing
number of large biomedical databases and electronic health
records are vital resources for healthcare researchers. However
recent works have shown that sharing this data when aggregated
to produce p-values, regression coefficients, count queries, and
Minor Allele Frequencies (MAFs) may cause compromise to patient
privacy [2].
The question is; can patient privacy be protected while still
making the most out of medical data? Extensive sharing and reuse
of medical data are usually endorsed by many, but participants often
expect that their privacy to be preserved. To achieve privacy, many
organizations are adopting various legal protections, such as Data
Use Agreements (DUAs) that explicitly eliminate re-identification
(Paltoo et al.) and technical controls, such as the suppression
or noise addition to genomic variants having a high likelihood of
distinguishing an individual [3]. However, reports have shown that
preservation of privacy might be impossible to realize, despite
sharing of only summary statistics [4]. There are also reports over the
past decade on how de-identified genomic data have been tracked
back to named persons, leading to public apologies and dramatic
policy changes (Shringarpure and Bustamante). Various genomic
statistics such as Minor Allele Frequency (MAF) and regression
coefficients can lead to privacy concerns [5]. This understanding
has led various groups removing statistical data from public
databases into access-controlled format. Though such protections
help preserve privacy, they also have adverse effects on access to
useful dataset for medical research. The medical community is at
a crossroad; how can researchers access medical data to data to
save lives and still ensure the privacy of the individuals involved
in the datasets. An effective model for genomic data dissemination
can be achieved through an approach based on game theory to
account for adversarial behaviors and capabilities. The proposed
approach has already been used to analyze the reidentification risk
and proven effective in some risk inherent domains, such as airport
security and coast guard patrols [6]. Methodologies are borrowed
from game theory to develop an effective, measurable protections
for genomic data sharing. This method accounts for adversarial
behavior to balance risks against utility more effectively compared
with traditional approaches.
Review of Related Works
There are many approaches in ensuring biomedical data privacy: Non-cryptographic and cryptographic approaches [7]. In this section, a brief summary of existing non-cryptographic techniques are presented.
Non-cryptographic approach
Non-cryptographic approaches adopt various sanitization
techniques to ensure the privacy of genomic data. Privacy
Preserving Data Publishing (PPDP) is a well-studied domain and
has been researched extensively for various types of data. These
techniques study how to transform raw data into a version that is
immunized against privacy attacks but that still preserves useful
information for data analysis. Existing techniques first sanitize raw
data and then release the sanitized data for public use. Once shared,
the data owner has no further control over the shared data. Existing
techniques are primarily based on two major privacy models: k
anonymity and ε-differential privacy. Despite its wide applicability
in the healthcare domain, recent research results indicate that
k anonymity-based techniques are vulnerable to an adversary’s
background knowledge [8]. This has inspired a discussion in the
research community in favor of the ε-differential privacy model,
which provides provable privacy guarantees independent of
an adversary’s background knowledge. However, it is not well
understood whether differential privacy is the right privacy model
for biomedical data as it fails to provide adequate data utility.
To satisfy a specific privacy model, while many anonymization
techniques have been proposed for various type of data; relational,
set-valued, spatio-temporal data, the problem of genomic data
anonymization has been little studied.
One of the limitations of the non-cryptographic approach
is that there is a trade-off between privacy and utility. All the
proposed methods compromise significant amount of utility
while protecting privacy. Differentially private mechanism may
provide wrong information due to noise addition. Therefore,
cryptographic approach has recently received much attention as
an alternative approach to protect genomic data privacy. [5] proved
that de-identification is an ineffective way to protect the privacy of
participants in genome-wide association studies, Recently, it has
been shown how they identified DNAs of several individuals (and
their families) who participated in scientific studies [9].
Several algorithms for inference on graphical models have
been proposed in the context of pedigree analysis. Exact inference
techniques on Bayesian networks are used in order to map disease
genes and construct genetic maps. Monte Carlo methods (Gibbs
sampling) were also proved to be efficient for genetic analyses in
the case of complex pedigrees (Sheehan). All these methods aim to
infer specific genotypes given phenotypes (like diseases). Another
paper relies on Gibbs sampling in order to infer haplotypes (used in
association studies) from genotype data [10]. Genotype imputation
is another technique used by geneticists to complete missing SNPs
based upon given genotyped data. A similar approach has recently
been used to infer high-density genotypes in pedigrees, by relying
notably on low-resolution genotypes and identity-by-descent
regions of the genome [11]. None of these contributions addresses
privacy. Johnson and Shmatikov proposed privacy-preserving
algorithms for computing various statistics related to the SNPs,
while guaranteeing differential privacy. However, differential
privacy reduces the accuracy of research results and is aimed to be
applied on aggregate results. In our work, we focus on protecting
individual genomic data. Some works also focus on protecting the
privacy of genomic data and on preserving utility in medical tests
such as
(i) searching of a particular pattern in the DNA sequence
(Troncoso et al.) and (Blanton and Aliasgari).
(ii) comparing the similarity of DNA sequences, [12].
(iii) performing statistical analysis on several DNA sequences [13].
[14] proposed privacy-preserving schemes for medical tests and
personalized medicine methods that use patients’ genomic data.
For privacy-preserving clinical genomics, a group of researchers
proposes to outsource some costly computations to a public cloud
or semi-trusted service provider.
Methodology
Preamble
This framework is designed to increase access to large-scale genomic data while promoting privacy using a game theoretic approach. Game models are built to formulate (formalize) the interactions among data owners and backward induction approach is proposed to find the Nash equilibria of the game. A model for genomic data dissemination and sharing is designed to account for adversarial behavior and capabilities. The model is used to demonstrate how a game theoretic approach can improve data sharing. Game theory is adapted for modeling interactions involved in genomic data sharing process to protect privacy. The decision of a data sharer is affected by several factors, including his personal privacy preference (e.g. whether he cares much about privacy), the incentives offered by the data collector, and the level of privacy protection that the data collector guarantees.
System Design Overview
Two actors are to play the game: SNP sharer who could be an
investigator of a study or an organization, such as an academic
medical center, and the recipient (or researcher), who would
request to access the data for some purpose (for example., research
purpose, findings or discovery of new associations).
The majority of recipients are unlikely to misuse the data, but
the privacy concern is on those with the potential to exploit named
genomes (or targets) by determining their presence in the dataset.
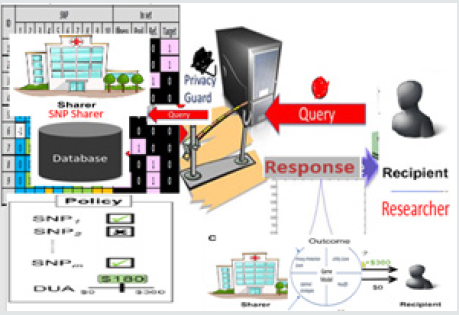
In this model, the sharer is a leader who can
a. require a DUA with liquidated damages in the event of a breach
of contract and
b. share a subset of SNP summary statistics from a specific study
(suppressing the rest).
The recipient of the data then follows by determining whether
the benefits gained by attacking each target outweigh the costs.
Crucially, the sharer chooses the policy that optimally balances
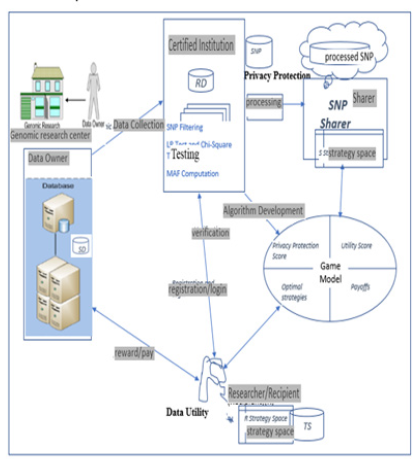
the anticipated utility and privacy risk. Figure 1 presents a general
architecture of the proposed framework. The game is played
between SNP sharer and researcher. As depicted in the Figure, it
incorporates four main participants: Data Owners (DOs), Certified
Institution (CI), SNP Sharer and Researchers (Recipients). The
functions performed by each of the entities are discussed below
(Figures 2 &3):
DOs consist of the institutions that agreed to share genomic data (that is, study dataset (SD)) they possess. These institutions might be any academic institutions, non-academic research organizations, government research agency or health departments. They collect samples from donors, carry out DNA sequencing and provide the formerly agreed digitized format to CI. CIs are the institution that have legal authority to process the raw dataset from DOs before sending it to SNP sharer. The data shared by different data owners reside in a database owned by the trusted entity. Any government institution such as National Institute of Health (NIH) in United States, Nigerian Institute of Medical Research (NIMR) and African Center of Excellence for Genomics of Infectious Diseases (ACEGID) in Nigeria can play this role. The main responsibilities performed by CI are: SNP Filtering, MAF Computation and Privacy Protection (PP). PP is done by checking data resistance to one of the strongest re-identification statistical attack (likelihood ratio test).
SNP Sharer (Publisher)
The SNP Sharer (also known as publishers) are biomedical researchers who are disseminating research datasets. Funding organizations, such as the NIH, NIHR and ACEGID require researchers who are granted funding to publish the data generated by their research through websites such as the Database of Genotypes and Phenotypes (dbGaP) .However, while they need to share data, they also have an incentive to protect the identities of the individuals who participated in the original research (that is, ensure Privacy of data) while recipient is interested in the data utility. The benefit associated with publishing the research dataset can be correlated to the amount of funding received for the project. For example, consider the dataset in dbGaP submitted by the five separate member institutions of the NIH-sponsored Electronic Medical Records and Genomics Network (EMERGE) [15].
Researchers (Recipient)
Researchers might be any individual or organization who is interested in executing query on the aggregate shared data residing in the CS. To execute query on the outsourced data, researchers need to log in with registered password which is stored CI database. The recipient is modeled as an intelligent attacker who can access external resources (called Target set) at a fixed cost to perform a reidentification attack, only attempts re-identification if his associated benefits exceed the costs (which can also include linking and curation costs) [16,17].
Conclusion
In this paper, a privacy-preserving technique for biomedical data using game theory has been proposed. The main contribution to data privacy research is the design of a model for representing major parties’ interactions (Data Owners, Certified Institution, SNP Sharer and Researchers (Recipients)) involved in genomic data sharing. This is to ensure that sharing and dissemination activities are captured in order to protect privacy.
References
- Akgun M, Bayrak AO, Ozer B, Sagiroglu MS (2015) Privacy preserving processing of genomic data: A survey. Journal of biomedical informatics 56: 103-111.
- Barth D, Khadam El E, Bambauer J, Cavoukian A, Malin B (2015) Assessing data intrusion threats. Science. Data Privacy Management International Workshop 348(6231): 194-195.
- Simmons S, Sahinalp C, Berger B (2016) Enabling privacy preserving GWASs in heterogeneous human populations. Cell systems 3(1): 54-61.
- Rodriguez LL, Brooks LD, Greenberg JH, Green ED (2013) The complexities of genomic identifiability. Science 339(6117): 275-276.
- Homer N, Szelinger S, Redman M, Duggan D, Tembe W, et al. (2008) Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genetics 4(8): e1000167.
- Pita J, Jain M, Ordónez F, Portway C, Tambe M, et al. (2009) Using game theory for Los Angeles airport security. AI magazine 30(1): 43
- Aziz MA, Ghasemi R, Noman M (2017) Privacy and Security in the Genomic Era. ACM Conference on Computer and Communications Security (CCS) 7(4): 56-77.
- Wang MN, Chen R (2015) Differentially private genome data dissemination through top-down specialization. BMC Medical Informatics and Decision Making 14(1): 20-34
- Gymrek M, Mc Guire AL, Golan D, Halperin E, Erlich Y (2013) Identifying personal genomes by surname inference. Science 339(6117): 321-324.
- Kirkpatrick B, Halperin E, Karp RM (2010) Haplotype inference in complex Journal of Computational Biology 17(3): 269-280.
- Burdick JT, Chen WM, Abecasis GR, Cheung VG (2006) In silico method for Privacy in the Electronic Society 12(10): 730-736.
- Kerschbaum F, Beck M, Schonfeld D (2014) Inference control for privacy-preserving genome matching. arXiv:1405.0205
- Kantarcioglu M, Jiang W, Liu Y, Malin B (2008) A cryptographic approach to securely share and query genomic sequences. IEEE Transactions on information technology in biomedicine 12(5): 606-617.
- Ayday E, Raisaro JL, Laren M, Jack PJ, Hubaux T (2013) Privacy preserving computation of disease risk by using genomic, clinical, and environmental data. Proceedings of USENIX Security Workshop on Health Information Technologies 3(6): 56-69.
- Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, et al. (2013) The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genetics in Medicine 15(10): 761-771.
- Humbert M, Ayday E, Hubaux JP, Telenti A (2014) Reconciling utility with privacy in genomics. In Proceedings of the 13th Workshop on Privacy in the Electronic Society, p. 11-20.
- Shringarpure SS, Bustamante CD (2015) Privacy risks from genomic data-sharing beacons. The American Journal of Human Genetics 97(5): 631-646.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...