
Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4609
Research Article(ISSN: 2637-4609) 
Fungi Lipases Homology Modeling and Molecular Docking with Fatty Acids and Tripalmitin of Palm Oil Effluent Volume 4 - Issue 2
Jeel Moya-Salazar1,2* Joaquín Vértiz-Osores3, Sandro Jibaja4, Román Acevedo-Espinola5, Roció Rupa2, Mitchell Alarcón-Díaz3, Miluska Vega-Guevara6 and Robert Cucho- Flores7
- 1Hospital Nacional Docente Madre Niño San Bartolomé, Peru
- 2Facultad de Ciencias y Filosofía, Universidad Peruana Cayetano Heredia, Peru
- 3Escuela de posgrado, Universidad César Vallejo, Peru
- 1Universidad Ricardo Palma, Peru
- 2Facultad de Ciencias de la Salud, Universidad Privada Norbert Wiener, Peru
- 3Escuela de Posgrado, Universidad Marcelino Champagnat, Peru
- 3Universidad Alas Peruanas, Peru
Received: January 25, 2019; Published: June 18, 2019
*Corresponding author: Jeel Moya-Salazar, Facultad de Ciencias y Filosofía, Universidad Peruana Cayetano Heredia, Hospital Nacional Docente Madre Niño San Bartolomé, Lima, Peru
DOI: 10.32474/AOICS.2019.04.000183
Abstract
Lipases are enzymes that catalyze the hydrolysis of long chain triglycerides. These enzymes have a key role in several human practices and industries, and it has been used in bioremediation processes. Due to the stability and specificity to their substrates under various conditions, their interest has grown during the last decade. The effluent from the palm oil industry contains about ~44% saturated fatty acids (mainly palmitic), ~37% monounsaturated fatty acids (mainly oleic acid) and 10% polyunsaturated fatty acids, potentially dangerous for the ecosystem. Bacteria and fungi contain lipases that are capable of degrade these lipids, but there are no sufficient studies on how these enzyme interacts with their substrates. In this study, by homology modeling, lipases 3D structure of Mucor circinelloides f. circinelloides 1006PhL and Rhizopus oryzae strains were modeled in silico using the SWISSMODEL server. The best models with stable structures were verified using the tool PROCHECK and Errat Server model. Further, a molecular docking was carried out between the selected modeled 3D structures with linoleic, linolenic, oleic, palmitic acid and tripalmitin using SwissDock tool to predict the lower binding energy of these lipases. Furthermore, the molecular docking analysis of 3D models of lipases from Mucor circinelloides f. circinelloides 1006PhL [EF405962] and Rhizopus oryzae [AER14043] suggest that pollutant fatty acids can bind to fungi lipases.
Keywords: Fatty acids; Fungi; Lipases; Homology modeling; Molecular docking
Introduction
Soil fungi are important for a wide variety of industrial processes. The use of fungi is based on their ability to produce enzymes of industrial interest, such as lipases that are widely used in the treatment of effluents. To date, the genera Mucor, Rhizopus, Penicillium, and Aspergillus have been tested with potential use in bioremediation [1-3].
Lipases (EC 3.1.1.3), a subclass of esterases, reversibly catalyze the hydrolysis of long-chain triglycerides [4]. It has recently been shown that fungal lipases display levels of activity and stability in non-aqueous environments. This remarkable feature facilitates esterification and transesterification [5]. Usually, lipid-specific reactions can be produced with high performance from fungi without requiring cofactors. In the case of bioremediation these enzymes have been used for degradation of palm oil effluent [1,6- 8].
In silico methods helps in the predictions of enzymatic affinity, activity, specificity, and selectivity of newly discovered proteins from structure information [9]. For example, molecular docking methods have also been applied successfully to predict most likely enzyme substrates with unknown experimental molecular interaction to its substrate [10]. This kind of bioinformatics approaches allow the previous screening of potential target for application in bioremediation and take advantage of fungi enzymes for industrial applications.
This method of predicting the enzyme-substrate complex structure can be accomplished through two unified steps: first, by sampling conformations of the ligand (e.g. fatty acids, tripalmitin, etc.) in the active site of the protein, and second by classifying these conformations by a score that ranks most negative value among all the conformations generated which indicates greater stability [11]. Molecular docking studies could help to identify pollutants fatty acids suitable to be degraded by these enzymes [12].
Although it has been reported amino acid sequences of lipases from bacteria and fungi with these properties tested in vitro [13], their crystal structure in Protein Data Bank has not yet been reported. Hence, our study is focused on predicting the 3D structure by homology modeling from lipase amino acids sequences of Mucor circinelloides f. circinelloides 1006PhL [EF405962] and Rhizopus oryzae [AER14043] and subsequently the interaction of modeled structures with fatty acids and tripalmitin were analyzed by molecular docking for calculating the full fitness energy (Kcal/mol).
Materials and Methods
Fungi sequence selection and alignment
The amino acid sequence of the reference lipase of Rhizopus microsporus var. Chinensis [EF405962] and the target amino acid sequences: Mucor circinelloides f. circinelloides 1006PhL [EPB86304] and R. oryzae [AER14043] were obtained from Genbank (http://www.ncbi.nlm.nih.gov/genbank/). We assessed whether all had registered their 3D structure UniProt (http:// www.uniprot.org/uniprot/) and PDB (http://www.rcsb.org/pdb/ home/home.do) databases. We inspected and adjusted manually the sequences using the software BioEdit v7.2.5 to diminish the number of gaps and insertions. Then, the alignment was performed with ClustalW2 and with PROSITE online tool (http://prosite. expasy.org) was determined the corresponding active site in PROSITE tool [14].
Homology modeling
The templates crystal structures were retrieved from RCSB Protein Data Bank (PDB) in base to identity (>30%). We did the homology modeling with SWISSMODEL (http://swissmodel.expasy. org/interactive), which is a fully automated protein structure homology-modeling server, accessible via the ExPASy web server [15]. The process considered the following steps:
(i) Template identification,
(ii) Template selection,
(iii) Model building and
(iv) Model quality estimation
The modeled structures were further verified with programs such as PROCHEK, which Checks stereochemical quality of protein structures [16] and Errat (http://www.doe- mbi.ucla.edu/ Services/Errat.html).
Molecular docking
Molecular docking was performed SwissDock server (http:// www.swissdock.ch/docking) to decipher the binding affinity and mode of interaction of the selected compounds (Scheme 1) against fungi lipases (of interest). SwissDock requires the target protein and ligand in PDB and Mol2 format, respectively. The ligands specified in Figure 1. As described by Grosdidier et al. [17], SwissDock is based on the docking software EADock DSS, whose algorithm consists of the following steps:
Figure 1: Chemical structures of ligands used for molecular docking. (A) Linoleic acid [PubChem: 4474613]; (B) linolenic acid [PubChem: 5280934]; (C) linolenic acid [PubChem: 3802189]; (D) Oleic acid [PubChem: 6845860]; (E) Palmitic acid [PubChem: 6072466]; and (F) Tripalmitin [PubChem: 8214701].
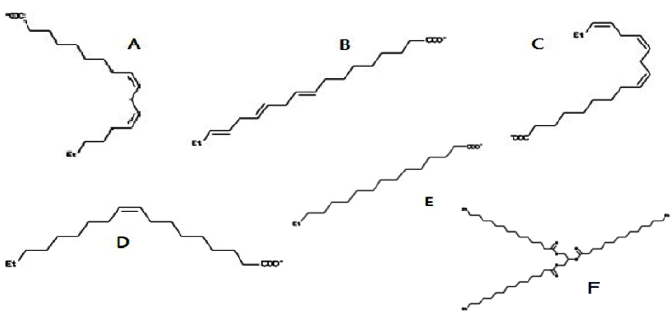
1. Binding modes are generated either in a box (local docking) or near all target cavities (blind docking).
2. Simultaneously, CHARMM energies of the binding modes are estimated on a grid.
3. The binding modes with most favorable energies are evaluated with FACTS, and clustered.
4. Most favorable clusters are visualized online and downloaded, and the obtained dock scores are reported in kcal/mol [17].
We analyzed docking results thru the UCSF Chimera program (RBVI, US), which provides an insight into the molecule to observe it specifically at the atomic level.
Results and Discussion
The amino acids sequences of the template and the query lipases were aligned with Clustal (Figure 2). The query sequences of Rhizopus oryzae [AER14043] and Mucor circinelloides f. circinelloides 1006PhL [EPB86304, M. circinelloides] consist of 294 and 360 residues, respectively, but the template structure was 389 residues. Manual editing query sequences were modeled from all its length and after alignment the sequence identity was 63.06% for Mucor circinelloides f. circinelloides 1006PhL [EPB86304] and 79.59% for Rhizopus oryzae [AER14043] respect to the sequence template Rhizopus microsporus var. Chinensis [EF405962]. We employed the result of alignment as criteria to build 3D structures for both query sequences by homology modeling.
Figure 2: Multiple alignments of amino acidic sequences of lipases from Rhizopus microsporus var. Chinensis [EF405962, Rh. microspoorus] and the target amino acid sequences: Mucor circinelloides f. circinelloides 1006PhL [EPB86304, M. circinelloides] and Rhizopus oryzae [AER14043, Rh. oryzae] performed with ClustalW2. Inside the box, the active site amino acids specific for lipases are shown (VI/VTGHSLGG).

Lipases are extensively distributed in prokaryotes and eukaryotes. Most conserved region in all these proteins focuses on a serine residue [18,19], with a residue of aspartic acid and a histidine in a charge relay system. Blow (1990) demonstrated their interaction and participation in a charge relay system [20]. Accordingly, with PROSITE our models have a serine active site VIVTGHSLGG specific for lipase, but Mucor circinelloides f. circinelloides 1006PhL [EPB86304] lipase model differs by one amino acid respect to the template protein (a valine replaced Isoleucine). The conserved histidine and aspartic acid residues must be highlighted in Figure 2.
Several studies suggest the broad and interesting use of the study with an in silico approach. Habitually they are used in the modern designs of drug to comprehend the mechanisms of pharmacological interaction at the molecular level, recognizing the network and the signaling pathways of genes and the substrateenzyme interactions. Due to the great development of the in silico studies, it is that the computational approaches can intensely support and help to decipher these specifics interactions between lipases and triglycerides [21]. The major objective of this study was to identify the binding energy of common pollutants fatty acids and tripalmitin present in palm oil effluents. Protein structure homology modeling has become a routine technique to generate 3D models for proteins when experimental structures are unavailable. The improved SWISS-MODEL pipeline makes extensive use of model quality estimation to select of most suitable templates and provides estimates of the expected accuracy of the resulting models [22].
A newly developed interactive web interface allows us to suitably search for appropriate templates using sensitive Hidden Markov Models searches against the SWISS-MODEL Template Library, analyses alternative templates and alignments, perform structural superposition and comparison, and compare the resulting models using mean force potential based model quality estimation tools. Model quality estimation is an essential part of protein structure predictions, as the accuracy of a model determines its usefulness for practical applications [22]. By, the structure of lipase from Mucor circinelloides f. circinelloides 1006PhL [EPB86304] and Rhizopus oryzae [AER14043] has not been crystallized yet; here the 3D structure of the lipase was predicted by homology modeling for both species favored by the high identity respect to its template.
We selected the best model among three 3D models generated after the verification of all parameters (Figure 3 & 4). Model quality declines with decreasing sequence identity. A typical model has ~1–2Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different [23]. Our Mucor circinelloides f. circinelloides 1006PhL [EPB86304] and Rhizopus oryzae [AER14043] models have 0.28Å and 0.07Å root mean square deviations between the matched Cα atoms with respect to template structure, respectively.
Figure 3: Errat plots of modeled lipases from Mucor circinelloides f. circinelloides 1006PhL (upper) [EPB86304], and Rhizopus oryzae (bottom) [AER14043]. The error values for model residues as predicted by ERRAT are shown. The “y” axis presents the error value and “x” axis presents the amino acid sequences of lipase. An error value exceeding 99% confidence level indicates poorly modeled regions.
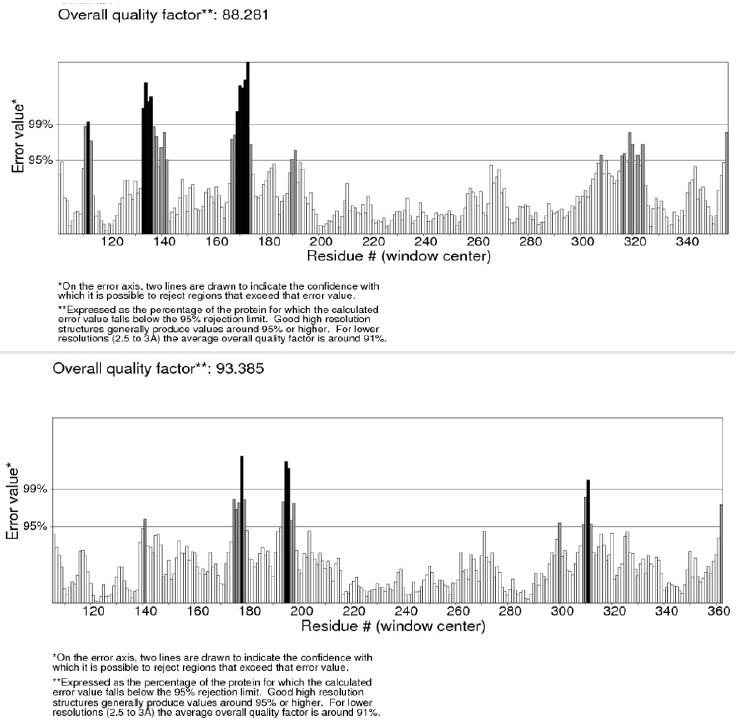
Figure 4: (A) The Ramachandran plot of modeled protein of Mucor circinelloides f. circinelloides 1006PhL [EPB86304] (left) showing 89.5% of the atom residing in the most favored region, 9.6% in the allowed region, 2.2% in the generously allowed region, and 0.4% residues was in the disallowed region. (B) The Ramachandran plot of modeled protein of Rhizopus oryzae [AER14043] (right) showing 89.7% of the atom residing in the most favored region, 8.6% in allowed region, 2.2% in generously allowed region, and 0.4% residues was in disallowed region.
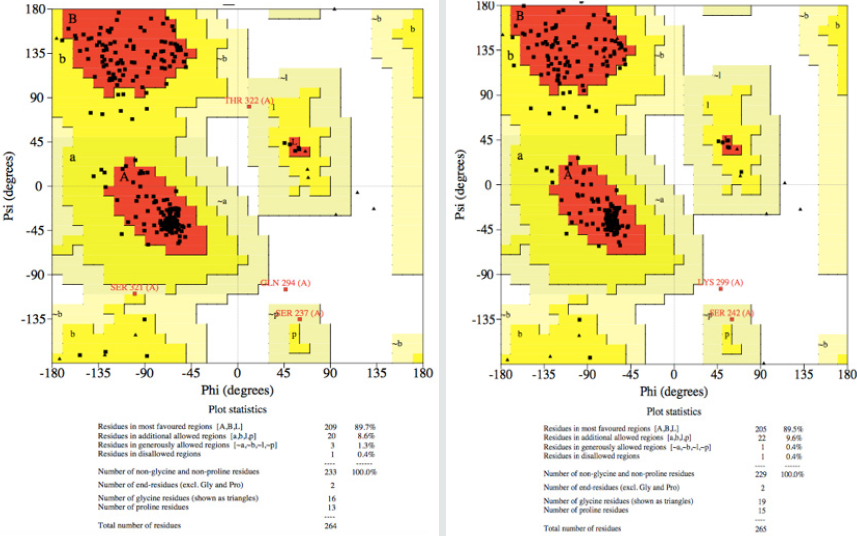
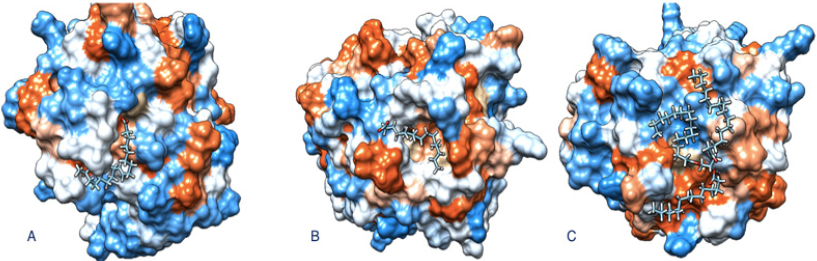
Figure 5: (A) Oleic acid molecule binds to Rhizopus oryzae modeled lipase 3D structure into the funnel form active site (-7.99, estimated ΔG). (B) Mucor circinelloides f. circinelloides 1006PhL modelled lipase, interacts with Palmitic acid (-6.45, estimated ΔG). (C) Tripalmitin binds to Rhizopus oryzae modeled lipase (-9.42, estimated ΔG).
The quality of the modeled structures was first analyzed with PROCHECK by Ramachandran plots. We would be expected to have a good quality model over 90% in most favored regions based on an analysis of a resolution of at least 2.0Å Angstroms and R-factor of ~20%.
The Ramachandran plot obtained from PROCHECK program with the Mucor circinelloides f. circinelloides 1006PhL [EPB86304] lipase model showed that 89.6% of residues were in most favored regions, 9.6% residues were in additional allowed regions, 1.3% in the generously allowed regions, and 0.4% in the disallowed regions. Likewise, in the same way we validated the Rhizopus oryzae [AER14043] lipase model with 89.7% of total residues in most favored regions, 8.6% residues in additional allowed regions, 0.4% in the generously allowed and disallowed regions (Figure 5).
We also used ERRAT (http://nihserver.mbi.ucla.edu), which analyses the statistics of non-bonded interactions between different atom types. A single output plot was produced by Errat program that gave the value of the error function against position of residues [24]. An error value exceeding 99% confidence level indicates poorly modeled regions (Figure 3). Errat analyses also provide an overall quality factor, expressed as the percentage of the protein for which the calculated error value falls below the 95% rejection limit. Good high resolution structures generally produce values around 95% or higher. For lower resolutions (2.5–3Å) the average overall quality factor is around 91% [24]. The overall quality factor assigned for Mucor circinelloides f. circinelloides 1006PhL [EPB86304] and Rhizopus oryzae [AER14043] lipase models were 88.3 and 93.4, respectively.
Accordingly, with docking study published here, it was predicted that modeled lipases have most effective interaction with tripalmitin (binding energy of -9.57Kcal/mol). Docking analysis of Mucor circinelloides f. circinelloides 1006PhL modeled lipase with fatty acids and tripalmitin was performed and full fitness energy was recorded. The results showed a good interaction more than one-fatty acids and tripalmiting them (Table 1). We performed the docking with SwissDock with rigid selection. We conducted the same procedure with Rhizopus oryzae modelled lipase with these substrates under the same parameters and software and the results were appropriate scores for binding.
Table 1: Molecular docking scores for target lipase structures and possible substrates found in palm oil.
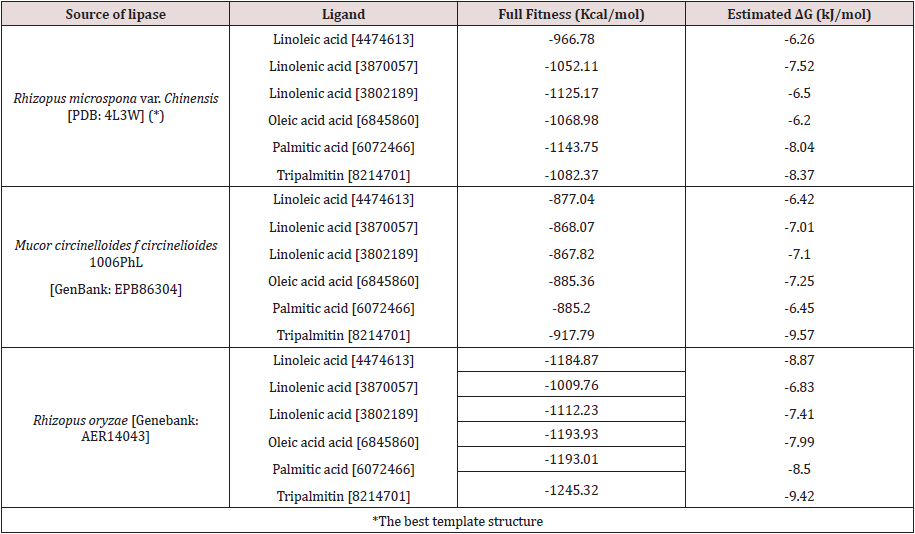
As a reference, a Rhizopus microsporus var. Chinensis [EF405962] lipase with PDB ID 4L3W was taken for comparison, where accordingly with previous reports [18,25-27]. We recognized that the triad Arg–Asp–Pro (RDP) is preserved in the lipase, responsible for the detection of the molecules tested. Our data revealed the above mentioned (VIVTGHSLGG) conserved domain in the lipase from Mucor circinelloides f. circinelloides 1006PhL [EPB86304] and Rhizopus oryzae [AER14043] as the main site for lipase activity. We also observed that fatty acids and tripalmitin binds into the pocket of the funnel shape active site of both lipases and bonding between the molecules is through hydrogen bond with bond length ~2.50Å, as previously reported with another ligand/enzyme [15]. In contrast, when we analyzed the result in the lipase the docking score was favorable and fatty acid and tripalmitin molecule were bind, given to the protein low fullfitness energy (Table 1).
Tang et al. [28], report that Valine plays a key role in stabilizing the accurate orientation of and maintaining the enantioselectivity of the lipase [28]. In comparison with our study Mucor circinelloides f. circinelloides 1006PhL [EPB86304] modeled lipase showed a substitution of isoleucine (I) to valine (V) in the active site (Figure 2). However, Rhizopus oryzae [AER14043] do not have this change and the modeled lipase showed slightly more favorable energy (more negative) for some substrates.
Undoubtedly, the new findings on the lipases’ structurefunction relationships are opening significant exploration ways for applications in several industries. For example, in industrial bioremediation, our findings on functional modification of enzymes will conduct to the rational design of proteins to perhaps improve their substrate specificity, enantioselectivity, catalytic efficiency and thermostability. Lately, Zhang et al. [29] were shown to improve the performance of Rhizomucor miehei lipase (RML) exhibited on yeast surface in the production of human milk fat substitute (HMFS) [29]. They developed an amino acid mutation in the lipase substratebinding pocket based on protein hydrophobicity, improving its activity.
They used molecular coupling to calculate the binding energy between lipase and substrates, finding that one mutant had significantly lower energy when oleic acid (-3.97KJ/mol) and tripalmitin (7.55KJ/mol decrease) were subs Zhang M., X.W. Yu, Y. Xu, et al. Crystal structure of lipase from Rhizopus microsporus var. chinensis. 2013; (en linea) [http://ftp.wwpdb.org/pub/pdb/ validation_reports/l3/4l3w/4l3w_full_validation.pdf]. Acceso 10/12/2017 trates. They used molecular docking to calculate the binding energy between lipase and substrates, finding that one mutant had significantly lower energy when oleic acid (-3.97KJ/ mol) and tripalmitin (7.55KJ/mol decrease) were substrates. However, although we did not change any residue, we obtained higher scores for oleic acid (~ 2.5 times for our modeled structures) and for tripalmitin ~ 9.5 for both modeled structures, suggesting tremendous future application in bioremediation for the informed lipases here.
Conclusion
The molecular docking analysis of 3D models of lipases from Mucor circinelloides f. circinelloides 1006PhL [EF405962] and Rhizopus oryzae [AER14043] suggest that pollutant fatty acids can bind to fungi lipases. The structural modeling study for Mucor circinelloides f. circinelloides 1006PhL [EPB86304] and Rhizopus oryzae [AER14043] lipase is reported in this study being Rhizopus oryzae [AER14043] the best model. This study highlights fungal lipases and exemplifies its binding to the substrate, i.e. palmitic, oleic acid, tripalmitin, even values more favorable than the template structure. However, these compounds are potential substrates for these lipases and we suggest that could be evaluated in vitro. This is the first study of molecular docking between fatty acids and a tripalmitin of palm oil showing an adequate binding energy accordingly to previous studies with other fungi lipases. Future molecular studies will be made for redesigned the models with specifics mutations for perhaps a better interaction with pollutant fatty acids and triglycerides for applications in industry.
References
- Mateos JC, Rodríguez JA, Roussos J, Cordova A, Abousalham F, et al. (2006) Lipase from the thermotolerant fungus Rhizopus homothallicus is more thermostable when produced using solid state fermentation than liquid fermentation procedures. Enzyme Microb Technol 39(5):1042–1050.
- Cruz-Ramírez MG, Rivera-Ríos JM, Téllez-Jurado A, Maqueda Gálvez AP, Mercado-Flores Y, et al. (2014) Screening for thermotolerant ligninolytic fungi with laccase, lipase, and protease activity isolated in Mexico. J Environ Biol 35(3): 521-529.
- Okogbenin OB, Anisiobi GE, Okogbenin EA, Ojieabu A (2014) Microbiological assessment and physiochemical parameters of palm oil mill effluent collected in a local mill in Ovia North East area of Edo State, Nigeria. Herald journal of Microbiology Biotechnology 1(1): 1–9.
- Lima ACP, Cammarota MC, Gutarra MLE (2018) Obtaining filamentous fungi and lipases from sewage treatment plant residue for fat degradation in anaerobic reactors. Peer J 6: e5368.
- Efimova E, Marjakangas JM, Lakaniemi AM, Koskinen PE, Puhakka JA (2013) Lipid profile characterization of wastewaters from different origins. Water Sci Technol 68(11): 2505-2514.
- Mendoza LI (2010) Aislamiento y selección de hongos lipolíticos a partir de aceites vegetales de desecho (proveniente de frituras) utilizados en la elaboración de biodiesel [Thesis]. Facultad de Ciencias biológicas, Universidad Nacional Mayor de San Marcos, Peru.
- Pruksatrakul T, Phoopraintra P, Wilairat P, Chaiyen P, Chantiwas R (2017) Development of a sequential injection-liquid microextraction procedure with GC-FID for analysis of short-chain fatty acids in palm oil mill effluent. Talanta 165: 612-618.
- Gopinath SC, Anbu P, Lakshmipriya T, Hilda A (2013) Strategies to characterize fungal lipases for applications in medicine and dairy industry. Biomed Res Int 2013: 154549.
- Singh RK, Feller A, Roovers M, Van Elder D, Wauters L, et al. (2018) Structural and biochemical analysis of the dual-specificity Trm10 enzyme from Thermococcus kodakaraensis prompts reconsideration of its catalytic mechanism. RNA 24(8):1080-1092.
- Vasel B, Hecht HJ, Schmid RD, Schomburg D (1993) 3D-structures of the lipase from Rhizomucor miehei at different temperatures and computer modelling of a complex of the lipase with trilaurylglycerol. J Biotechnol 28(1): 99-115.
- Meng XY, Zhang HK, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7(2): 146-157.
- Ericsson DJ, Kasrayan A, Johansson P, Bergfors T, Sandström AG, et al. (2008) X-ray structure of Candida antarctica lipase A shows a novel lid structure and a likely mode of interfacial activation. J Mol Biol 376(1): 109-119.
- Nwuche CO, Ogbonna JC (2011) Isolation of lipase producing fungi from palm oil mil eflfuent (POME) dumps sites al Nsukka. Braz Arch Boil Technol 54(1): 113-116.
- Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, et al. (2010) PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38: D161–D166.
- Singh PK, Shukla P (2012) Molecular modeling and docking of microbial inulinases towards perceptive enzyme-substrate interactions. Indian J Microbiol 52(3): 373-380.
- Morris AL, MacArthur MW, Hutchinson EG, Thornton JM (1992) Stereochemical quality of protein structure coordinates. Proteins 12(4): 345-364.
- Grosdidier A, Zoete V, Michielin O (2011) Swiss Dock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 39: W270-W277.
- Rehm S, Trodler P, Pleiss J (2010) Solvent-induced lid opening in lipases: a molecular dynamics study. Protein Sci 19(11): 2122-2130.
- Borrelli GM, Trono D (2015) Recombinant Lipases and Phospholipases and Their Use as Biocatalysts for Industrial Applications. Int J Mol Sci 16(9): 20774-20840.
- Blow D (1990) Enzymology. More of the catalytic triad. Nature 343: 694-695.
- Juhl PB, Trodler P, Tyagi S, Pleiss J (2009) Modelling substrate specificity and enantioselectivity for lipases and esterases by substrate-imprinted docking. BMC Struct Biol 9: 39.
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252-W258.
- Grosdidier A, Zoete V, Michielin O (2011) Fast docking using the CHARMM force field with EADock DSS. J Comput Chem 32(10): 2149-2159.
- Sehgal SA, Tahir RA, Mirza AH (2018) Quick Guideline for Computational Drug Design. Bentham eBooks, Netherlands.
- Zhang JH, Jiang YY, Lin Y, Sun YF, Zheng SP, et al. (2013) Structure-guided modification of Rhizomucor miehei lipase for production of structured lipids. PLoS One 8(7): e67892.
- Yang S, Qin Z, Duan X, Yan Q, Jiang Z (2015) Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei. J Lipids Res 56(8): 1616-1624.
- Zan X, Tang X, Chu L, Zhao L, Chen H, et al. (2016) Lipase genes in Mucor circinelloides: identification, sub-cellular location, phylogenetic analysis and expression profiling during growth and lipid accumulation. J Ind Microbiol Biotechnol 43(10):1467-1480.
- Tang L, Su M, Chi L, Zhang J, Zhang H, et al. (2014) Residue Val237 is critical for the enantioselectivity of Penicillium expansum Biotechnol Lett 36(3): 633-639.
- Zhang M, Yu XW, Xu Y, Huang CH, Guo RT (2013) Crystal structure of lipase from Rhizopus microsporus var. chinensis.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...





