Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-5945
Review Article(ISSN: 2638-5945) 
On the Neglected Shifting Balance Theory, Bateson– Dobzhansky–Muller Model & Quantum Evolution Plus the Role of Mitochondrial Membrane Potential (Mmp) Impact on Covid-19 Volume 4 - Issue 2
Sorush Niknamian*
- Military Medicine, Liberty University (LU), VA, USA, Fellow Member of International Society of Infectious Disease (ISID)
Received: February 19, 2021 Published: March 03, 2021
Corresponding author: Sorush Niknamian, Military Medicine, Liberty University (LU), Fellow Member of International Society of Infectious Disease, USA
DOI: 10.32474/OAJOM.2021.04.000186
Abstract
Background: Approximately 80% of all viruses are RNA viruses and they contain their specific RNA helicases. Defective RNA helicases have been linked to infectious diseases (Viral Infections). Materials and Methods: The articles have gone through many types of research from the beginning of the epidemic of Coronaviruses through history and we introduced the neglected hypothesis of Shifting balance theory, Bateson–Dobzhansky–Muller model & Quantum evolution. In the ancestral population, the genotype is AABB. When two populations become isolated from each other, new mutations can arise. In one population A evolves into a, and in the other B evolves into b. When the two populations hybridize it is the first time A and B interact with each other. When these alleles are incompatible, we speak of Dobzhansky–Muller incompatibilities plus the role of MMA in mitochondria in spreading SARS-CoV-19 through populations and the result of an infection in COVID-19.
Results: In viruses specifically COVID-19, Ribosomal Frameshift is programmed to allows the virus to encode multiple types of proteins from the same mRNA. HIV-1 (human immunodeficiency virus), RSV (Rous sarcoma virus), and all types of influenza viruses use Ribosomal Frameshift. they rely on frameshifting to create a proper ratio of normal translation and trans-frame (encoded by frameshifted sequence) proteins. Notably, its use in viruses is primarily for compacting more genetic information into a shorter amount of genetic material.
Conclusion: to find the genome sequence of COVID-19 we also used Nanopore sequencing that introduced and manufactured by Oxford scientists, due to differences in the action of infection in the host, we could not reach any results since the Novel Virus has not a stable genome (which is quite dynamic) since through our deep research, each virus contains its specific genome sequencing and we cannot claim that COVID-19 has one specific genome sequence like MERS-CoV, SARS-CoV or any types of viruses which has been discovered and contains their specific genome.
Keywords: Shifting Balance Theory; COVID-19; MMP; Bateson-Dobzhansky-Muller Model & Quantum Evolution
Introduction
Coronaviruses were discovered in the 1930s [1]. Arthur Schalk and M.C. Hawn figure out in 1931 a new respiratory infection (og Coronaviruses) in North Dakota. The infected chickens’ mortality rate was 40%–90% [2]. Fred Beaudette and Charles Hudson isolated and cultivated the infectious bronchitis virus which caused the disease [3]. In the 1940s, during the world war, two more animal coronaviruses, mouse hepatitis virus (MHV), and transmissible gastroenteritis virus (TGEV) were isolated [4]. It was not realized at the time that these three different viruses were related to coronavirus [7]. Impotently, human coronaviruses were discovered in the 1960s [5-6]. They were isolated using two different methods in the UK and the US [8]. E.C. Kendall, Malcom Byone and David Tyrrell in 1960 isolated from a boy a new common cold virus B814 [9-11]. The virus, unfortunately, was not able to be cultivated using standard techniques which had successfully cultivated rhinoviruses, adenoviruses, and other known common cold viruses. In 1965, Tyrrell and Byone [12] The new cultivating method was introduced to the lab by Bertil Hoorn [13]. The isolated virus when intranasally inoculated into volunteers caused a cold and was inactivated by ether which indicated it had a lipid envelope [14-15]. Around the same time, Dorothy Hamre [16] and John Procknow at the University of Chicago isolated 229E virus from medical students, which they grew in kidney tissue culture. The novel virus 229E, like the virus strain B814, when inoculated into volunteers caused a cold and was inactivated by ether [17]. B814 and 229E viruses were subsequently imaged by electron microscopy in 1967 by Scottish virologist June Almeida at St. Thomas Hospital in London [18-19]. Not only B814 and 229E viruses where they related to each other, but they were morphologically related to infectious bronchitis virus (IBV) [20]. A research group at the National Institute of Health the same year was able to isolate another member of this new group of viruses using organ culture and named the virus strain OC43 (OC for organ culture) [21]. Like B814, 229E, and IBV, the novel cold virus OC43 had distinctive club-like spikes when observed with the electron microscope [22-23]. The IBV-like novel cold viruses were shown to be morphologically related to the mouse hepatitis virus [24]. This new group of IBV-like viruses came to be known as coronaviruses after their distinctive morphological appearance [25]. Human coronavirus 229E and OC43 continued to be studied for decades [26-27]. The coronavirus strain B814 was lost. It is not known which present human coronavirus it was [28]. Other human coronaviruses discovered and named SARS-CoV in 2003, HCoV NL63 in 2004, HCoV HKU1 in 2005, MERS-CoV in 2012, and SARSCoV- 2 in 2019 (COVID-19). [29-30]. There have also been many animal coronaviruses identified since the 1960s which shows these strains of viruses are high in number and very difficult to be studied one by one. [31].
Origin
The most recent common ancestor (MRCA) of all coronaviruses
is estimated to have existed as recently as 8000 BCE, although some
models place the common ancestor as far back as 55 million years
or more, implying long term coevolution with bat and avian species.
[32]. The most recent common ancestor of the alpha coronavirus
line has been placed at about 2400 BCE, of the beta coronavirus
line at 3300 BCE, of the gamma coronavirus line at 2800 BCE, and
of the delta coronavirus line at about 3000 BCE. Bats and birds, as
warm-blooded flying vertebrates, are an ideal natural reservoir
for the coronavirus gene pool (with bats the reservoir for alpha
coronaviruses and beta coronavirus- and birds the reservoir for
gamma coronaviruses and delta coronaviruses). The large number
and global range of bat and avian species that host viruses have
enabled extensive evolution and dissemination of coronaviruses.
[33].
Many human coronaviruses have their origin in bats [34].
The human coronavirus NL63 shared a common ancestor with a
bat coronavirus (ARCoV 2) between 1190 and 1449 CE [35]. The
human coronavirus 229E shared a common ancestor with a bat
coronavirus (GhanaGrp1 Bt CoV) between 1686 and 1800 CE
[36]. More recently, alpaca coronavirus and human coronavirus
229E diverged sometime before 1960 [68]. MERS-CoV emerged in
humans from bats through the intermediate host of camels. [37].
MERS-CoV, although related to several bat coronavirus species,
appears to have diverged from these several centuries ago.[38].
The most closely related bat coronavirus and SARS-CoV diverged
in 1986 [39]. A possible path of evolution of SARS coronavirus
and keen bat coronaviruses is that SARS-related coronaviruses
coevolved in bats for a long time. The ancestors of SARS-CoV first
infected leaf-nose bats of the genus Hipposideridae; subsequently,
they spread to horseshoe bats in the species Rhinolophidae, then to
civets, and finally to humans [40-41].
Unlike other Beta coronaviruses, bovine coronavirus of the
species Beta coronavirus 1 and subgenus Arbovirus is thought to
have originated in rodents and not in bats. [42-43]. In the 1790s,
equine coronavirus diverged from the bovine coronavirus after
a cross-species jump [44] (Table 1). Later in the 1890s, human
coronavirus OC43 diverged from bovine coronavirus after another
cross-species spillover event [45-46]. It is speculated that the flu
pandemic of 1890 may have been caused by this spillover event,
and not by the influenza virus, because of the related timing,
neurological symptoms, and unknown causative agent of the
pandemic [47]. Besides causing respiratory infections, human
coronavirus OC43 is also suspected of playing a role in neurological
diseases [48]. In the 1950s, the human coronavirus OC43 began to
diverge into its present genotypes [49]. Phylogenetically, mouse
hepatitis virus (Murine coronavirus), which infects the mouse’s liver
and central nervous system, [50]. is related to human coronavirus
OC43 and bovine coronavirus. Human coronavirus HKU1, like the
viruses, also has its origins in rodents [51]. Well-known rodents
include mice, rats, squirrels, prairie dogs, chipmunks, chinchillas,
porcupines, beavers, guinea pigs, hamsters, gerbils, and capybaras.
Rabbits, hares, and pikas (Figure 1).
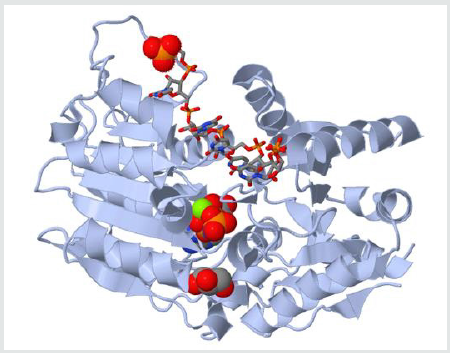
Figure 1: The different promoter sequences and accessory domains that aid in RNA unwinding (local strand separation). The regions in red are ATP binding domains and the regions in yellow are RNA interaction domains. Specific sequences termed DEAD-box proteins are also present that help catalyzes reactions in which ATP does not need to be directly hydrolyzed if it binds to the domains on the strand.
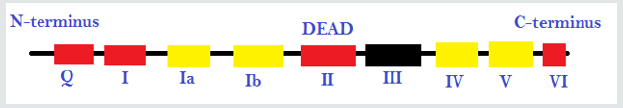
Materials and Methods
SARS-CoV-2 is a complex virus. the genome of the virus differs from one person to another like fingerprints and the reason behind this phenomenon is the amount of tRNA and the metabolism (Mitochondrial Function) of the individual which is based on the amount of ATP releasing from the Mitochondria of each cell and loss of MMP (mitochondrial membrane permeabilization) which the reason behind the cause of cell apoptosis. Mitochondrial ATP generation requires proteins from the nuclear and mitochondrial genomes. ROS (Reactive Oxygen Species) disrupt the oxidative production of ATP, which is required for normal cellular function, due to the damage of mtDNA disrupts the normal synthesis of proteins needed for mitochondria function and making them suitable targets for attack by ROS produced during infections by viruses especially, any virus including Coronaviruses raises ROS in the host cell which affects Mitochondria and leads to loss of MMP [52] (Figure 2).
Proteins are translated by reading trinucleotides on the mRNA strand, also known as codons, from one end of the mRNA to the other (from the 5’ to the 3’ end). Each codon is translated into a single amino acid. Therefore, a shift of any number of nucleotides that is not divisible by 3 in the reading frame will result in subsequent codons to be read differently [53]. This effectively changes the ribosomal reading frame. In viruses specifically COVID-19, Ribosomal Frameshift is programmed to allows the virus to encode multiple types of proteins from the same mRNA. HIV-1 (human immunodeficiency virus), [54] RSV (Rous sarcoma virus) [55], and all types of influenza viruses use Ribosomal Frameshift. they rely on frameshifting to create a proper ratio of normal translation and trans-frame (encoded by frameshifted sequence) proteins. Notably, its use in viruses is primarily for compacting more genetic information into a shorter amount of genetic material (Figure 3).
Effect of RNA Helicase in COVID-19
Helicases are enzymes that are vital to all living organisms.
Their main function is to unpack an organism’s genes. They
are motor proteins that move directionally along a nucleic acid
phosphodiester backbone, separating two annealed nucleic acid
strands such as DNA and RNA using energy from ATP hydrolysis
which proves that the vital COVID-19 is dependent on the amount
of ATP production of the host cell by mitochondria. Approximately
1% of eukaryotic genes code for helicases [56]. The human genome
codes for 95 non-redundant helicases: 64 RNA helicases (Which
is important in infection of SARS-CoV-2) and 31 DNA helicases.
Many cellular processes, such as DNA replication, transcription,
translation, recombination, DNA repair, and ribosome biogenesis
involve the separation of nucleic acid strands that necessitates
the use of helicases. This image represents the different promoter
sequences and accessory domains that aid in RNA unwinding (local
strand separation). The regions in red are ATP binding domains
and the regions in yellow are RNA interaction domains. Specific
sequences termed DEAD-box proteins are also present that help
catalyzes reactions in which ATP does not need to be directly
hydrolyzed if it binds to the domains on the strand.
Approximately 80% of all viruses are RNA viruses and they
contain their specific RNA helicases [58]. Defective RNA helicases
have been linked to infectious diseases (Viral Infections) [59].
Some RNA helicases and DNA helicases can be found together
in all the helicase super-families except for SF6 [60] [61]. All the
eukaryotic RNA helicases that have been identified up to date are
non-ring forming and are part of SF1 and SF2. On the other hand,
ring-forming RNA helicases have been found in bacteria and viruses
[62]. RNA helicases that do exhibit unwinding activity have been
characterized by at least two different mechanisms: canonical
duplex unwinding and local strand separation. local strand
separation occurs by a process wherein the helicase enzyme is
loaded at any place along with the duplex. This is usually aided by
a single-strand region of the RNA, and the loading of the enzyme is
accompanied by ATP binding [60]. Once the helicase and ATP are
bound, local strand separation occurs, which requires the binding
of ATP but not the actual process of ATP hydrolysis [62]. Presented
with fewer base pairs the duplex then dissociates without further
assistance from the enzyme. This mode of unwinding is used by the
DEAD/DEAH box helicases [63].
In Vitro, to find the specific helicase activity of COVID-19, we
used fluorescence-based assays, filtration assays, a scintillation
proximity assay, a time-resolved fluorescence resonance energy
transfer assay, and even used Tripoint diagnostic assay to
observe the Helicase Activity. Even we have used the basic strand
displacement assay which had been used in 1982-1983 [67,68].
The result was interestingly showed each Virus Helicase Assay is
different from the other. The reason is the environment where the
virus exits/evolves promptly and the only answer to this result is
Lamarckian Evolution. As Coronaviruses (Including SarS-CoV-2)
evolves very fast, their adaptation to the environment explains our
results [67-69].
Shifting Balance Theory, Bateson-Dobzhansky-Muller Model & Quantum Evolution
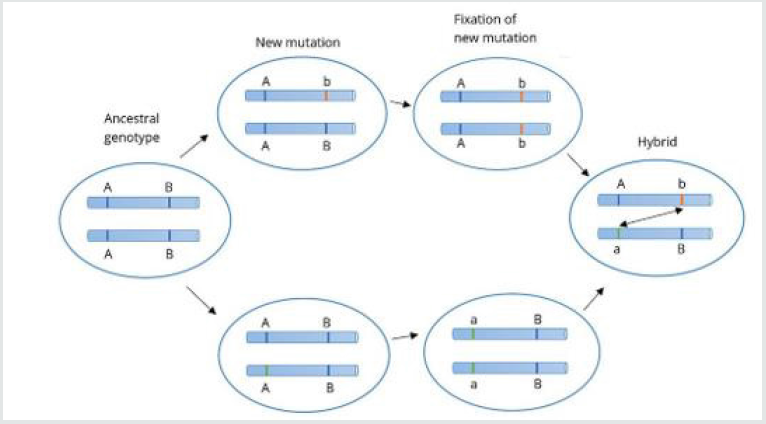
The Bateson–Dobzhansky–Muller model, [69]. is a model of the evolution of genetic incompatibility, important in understanding the evolution of reproductive isolation during speciation and the role of natural selection in bringing it about. The theory was first described by William Bateson [70] then by Theodosius Dobzhansky in 1934, and later by Herman Muller, et al. [71]. This model describes the drift between two species or even viruses to become hybrid and act differently in the environment. As many scientists only focus on the Co-Evolution of the viruses, Bateson- Dobzhansky-Muller model can be useful in describing the Fixation and Adaptation of SARS-CoV-2 in the different environment as well. In the ancestral population, the genotype is AABB. When two populations become isolated from each other, new mutations can arise. In one population A evolves into a, and in the other B evolves into b. When the two populations hybridize it is the first-time a and b interact with each other. When these alleles are incompatible, we speak of Dobzhansky-Muller incompatibilities. The Shifting balance theory is another theory of evolution proposed in 1932 by Sewall Wright, suggesting that adaptive evolution may proceed most quickly when a population of viruses divides into subpopulations with restricted gene flow. attempting to explain how a population may move across an adaptive valley to a higher adaptive peak [72] (Figure 4).
Figure 4: The arrows indicate the preferred flow of a population (Viruses as well) on the landscape.
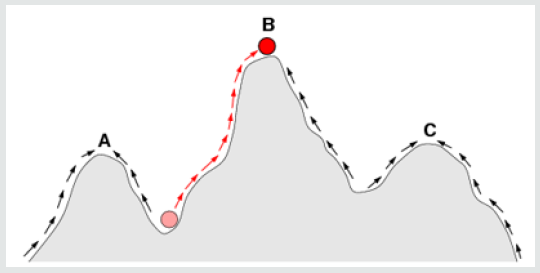
According to the theory, this movement occurs in three steps:
a) Genetic drift allows a locally adapted subpopulation to
move across an adaptive valley to the base of a higher adaptive
peak.
b) Natural selection will move the subpopulation up to the
higher peak.
c) This new superiorly adapted subpopulation may then
expand its range and outcompete or interbreed with other
subpopulations, causing the spread of new adaptations and
movement of the global population toward the new fitness peak.
All three steps describe adaption, Genetic Drift, and the Fitness
of SARS-CoV-2. Novel Coronavirus has been evolved so fast from
their ancestors to become a new Hybrid Novel RNA Virus and like
cancer cells, they have rapid genetic mutations and adaptation to
the environment.
The red ball indicates a population (or Viruses) that moves from
an adaptive valley to the top of an adaptive peak. Based on natural
selection (which usually acts to increase fitness in a population), it is
not possible for a population at peak A to reach peak B because this
requires descending into an adaptive valley. Shifting balance theory
aims to explain how this may be possible. Quantum evolution was
proposed by George Gaylord Simpson in 1953. Quantum Evolution
happens at the Taxonomic level and it plays a major role in the
origin taxonomic units of relatively high rank in families, orders,
and classes of species and parasites including viruses. According
to Simpson’s statements in 1944, quantum evolution resulted from
Sewall Wright’s model of random genetic drift [74-83].
The History Behind the Neglected Coronavirus
We cannot forget the history of the outbreak of Coronaviruses. Firstly, it was considered harmless pathogens until they caused three major outbreaks of severe respiratory disease in the last 20 years. The Coronavirus was recognized in 1960 [84] and it was identified as a cause of the common cold. In 2002, it was considered as a nonfatal virus not severely pathogenic to humans. Then took place across the globe when an infected doctor traveled to Hong Kong in February 2003 [85] and transmitted the infection to other health workers and guests staying in the same hotel. These patients brought infection back to their home countries, that is, Singapore, Vietnam, and Canada. In the year 2012 [86], a new coronavirus emerged in the Middle East and was named Middle East Respiratory Syndrome (MERS) [86-90]. Through our sessions above, the COVID-19 is a Hybrid Virus with a high mutation rate like cancer cells and has the potential of causing a new Hybrid Pandemic in the future. The Shifting Theory, Co-Evolution, Bateson–Dobzhansky- Muller model, and Quantum Evolution are the result of these high rare pandemics in the last 20 years.
Conclusion
Based on our research, to find the genome sequence of COVID-19 we also used Nanopore sequencing that introduced and manufactured by Oxford scientists, due to differences in the action of infection in the host, we could not reach any results since the Novel Virus has not a stable genome (which is quite dynamic) since through our deep research, each virus contains its specific genome sequencing and we cannot claim that COVID-19 has one specific genome sequence like MERS-CoV, SARS-C-V or any types of viruses which has been discovered and contains their specific genome. The main reason is the quick adaptation to the environment, Temperature, humidity, host genome type, host metabolism, Genetic Drift, Recombination of the virus, the high population of human beings, and the amount of ATP production of the host by their Mitochondria. Therefore, any type of vaccine cannot prevent the host from becoming infected by the virus. Vice versa, the new Coronavirus becomes more adapted and changes rapidly to make the environment and the host becoming weak and infected in the end.
References
- Estola T (1970) Coronaviruses, a New Group of Animal RNA Viruses. Avian Diseases 14(2): 330-336.
- Fabricant J (1998) The Early History of Infectious Bronchitis. Avian Diseases 42(4): 648-650.
- Decaro N (2011) Gammacoronavirus. In Tidona C, Darai G (eds.) Gammacoronavirus‡: Coronaviridae. The Springer Index of Viruses Springer 403-413.
- McIntosh K (1974) Coronaviruses: A Comparative Review". In Arber W, Haas R, Henle W, Hofschneider PH, Jerne NK, Koldovský P, Koprowski H, Maaløe O, Rott R (eds.) Current Topics in Microbiology and Immunology / Ergebnisse der Mikrobiologie und Immunitätsforschung. Current Topics in Microbiology and Immunology/Ergebnisse der Mikrobiologie And Immunitätsforschung. Berlin, Heidelberg: Springer p. 87.
- Il était une fois les coronavirus. Réalités Biomédicales (in French). 2020-03-27. Retrieved 2020-04-18.
- Kahn JS, McIntosh K (2005) History and recent advances in coronavirus discovery". The Pediatric Infectious Disease 24(11 Suppl): S223-227, discussion S226.
- Mahase E (2020) The BMJ in 1965. BMJ 369: m1547.
- Monto AS (1984) Coronaviruses". In Evans AS (ed.). Viral Infections of Humans. Viral Infections of Humans: Epidemiology and Control. Springer US. pp. 151-165. doi:10.1007/978-1-4684-4727-9_7. ISBN 978-1-4684-4727-9.
- Kendall EJ, Bynoe ML, Tyrrell DA (1962) Virus isolations from common colds occurring in a residential school". British Medical Journal. 2(5297): 82-86.
- Richmond C (2005) David Tyrrell BMJ: British Medical Journal 330 (7505): 1451.
- Obituary Notices: Malcom Byone (1969) British Medical Journal 2(5660): 827-829.
- Tyrrell DA, Bynoe ML (1965) Cultivation of a Novel Type of Common-Cold Virus in Organ Cultures". British Medical Journal 1(5448): 1467-1470.
- Tyrrell DA, Fielder M (2003) Cold Wars: The Fight Against the Common Cold. Oxford University Press 26(1): 93-95.
- Hagan WA, Bruner DW, Gillespie JH, Timoney JF, Scott FW, et al. (1988) Hagan and Bruner's Microbiology and Infectious Diseases of Domestic Animals: Regarding Etiology, Epizootiology, Pathogenesis, Immunity, Diagnosis, and Antimicrobial Susceptibility. Cornell University Press 440.
- Knapp, Alex (2020) The Secret History of The First Coronavirus. Forbes
- Hamre D, Procknow JJ (1966) A new virus isolated from the human respiratory tract". Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine. 121(1): 190-193.
- Dr June Almeida (2020) The woman who discovered the first coronavirus
- Almeida J (2008) June Almeida (née Hart). BMJ 336 (7659): PP.1511.
- Almeida JD, Tyrrell DA (1967) The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture". The Journal of General Virology. 1(2): 175-178.
- McIntosh K, Becker WB, Chanock RM (1967) Growth in the suckling-mouse brain of "IBV-like" viruses from patients with upper respiratory tract disease". Proceedings of the National Academy of Sciences of the United States of America. 58(6): 2268-2273.
- McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM, et al. (1967) Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease". Proceedings of the National Academy of Sciences of the United States of America 57(4): 933-940.
- Times, Harold M (1967) Six Newly Discovered Viruses May Explain Cold; Strains Are Similar to Germ That Causes a Bronchial Infection in Chickens Believed to Be New Group. The New York Times Schmeck Jr Special to the New York
- Myint SH (1995) Human Coronavirus Infections. In Siddell SG (eds.) The Coronaviridae The Viruses Springer US. 389-401.
- Geller C, Varbanov M, Duval RE (2012) Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses 4(11): 3044-3068.
- Corman VM, Jores J, Meyer B, Younan M, Liljander A, et al. (2014) Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013". Emerging Infectious Diseases. 20(8): 1319-1322.
- Su S, Wong G, Shi W, Liu J, Lai AC, et al. (2016) Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses". Trends in Microbiology 24 (6): 490–502.
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019". The New England Journal of Medicine. 382(8): 727-733.
- International Committee on Taxonomy of Viruses (2010-08-24). "ICTV Master Species List 2009-v10" (Xls).
- Cherry James, Demmler-Harrison, Gail J Kaplan, Sheldon L, Steinbach William J, et al. (2017) Feigin and Cherry's Textbook of Pediatric Infectious Diseases. Elsevier Health Sciences 2: p. PT6615.
- Chapter 24-Coronaviridae (2017) Fenner's Veterinary Virology (Fifth eds.). Academic Press pp. 435-461.
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJ, Talbot PJ, Woo PC, Ziebuhr J (2011) Family Coronaviridae". In King AM, Lefkowitz E, Adams MJ, Carstens EB, International Committee on Taxonomy of Viruses, International Union of Microbiological Societies. Virology Division (eds.). Ninth Report of the International Committee on Taxonomy of Viruses. Oxford: Elsevier. pp. 806-828.
- Wertheim JO, Chu DK, Peiris JS, Kosakovsky Pond SL, Poon LL, et al. (2013) A case for the ancient origin of coronaviruses". Journal of Virology 87(12): 7039-7045.
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, et al. (April 2012) Discovery of seven novels Mammalian and avian coronaviruses in the genus delta coronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and delta coronavirus. Journal of Virology 86(7): 3995-4008.
- Forni D, Cagliani R, Clerici M, Sironi M (2017) Molecular Evolution of Human Coronavirus Genomes". Trends in Microbiology 25(1): 35-48.
- Huynh J, Li S, Yount B, Smith A, Sturges L, et al. (2012) Evidence supporting a zoonotic origin of human coronavirus strain NL63. Journal of Virology 86(23): 12816-12825.
- Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, et al. (2009) Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerging Infectious Diseases 15 (9): 1377-1384.
- Crossley BM, Mock RE, Callison SA, Hietala SK (2012) Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E. Viruses. 4 (12): 3689-3700.
- Forni D, Cagliani R, Clerici M, Sironi M (2017) Molecular Evolution of Human Coronavirus Genomes. Trends in Microbiology 25 (1): 35-48.
- Lau SK, Li KS, Tsang AK, Lam CS, Ahmed S, et al. (2013) Genetic characterization of Beta coronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. Journal of Virology 87(15): 8638-8650.
- Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, et al. (2007) Evolutionary insights into the ecology of coronaviruses. Journal of Virology 81(15): 8371.
- Gouilh MA, Puechmaille SJ, Gonzalez JP, Teeling E, Kittayapong P, et al. (2011) SARS-Coronavirus ancestor's footprints in South-East Asian bat colonies and the refuge theory. Infection, Genetics, and Evolution 11(7): 1690-1702.
- Cui J, Han N, Streicker D, Li G, Tang X, Shi Z, et al. (2007) Evolutionary relationships between bat coronaviruses and their hosts. Emerging Infectious Diseases 13(10): 1526-1532.
- Lau SK, Woo PC, Li KS, Tsang AK, Fan RY, et al. (2015) Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. Journal of Virology. 89 (6): 3076-3092.
- Bidokhti MR, Tråvén M, Krishna NK, Munir M, Belák S, et al. (2013) Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains". The Journal of General Virology 94 (Pt 9): 2036-2049.
- Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, et al. (2005) Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. Journal of Virology 79(3): 1595-1604.
- Corman VM, Muth D, Niemeyer D, Drosten C (2018) Hosts and Sources of Endemic Human Coronaviruses. Advances in Virus Research 100: 163-188.
- Lau SK, Lee P, Tsang AK, Yip CC, Tse H, et al. (2011) Molecular epidemiology of human coronavirus OC43 reveals the evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. Journal of Virology 85(21): 11325-11337.
- Schaumburg CS, Held KS, Lane TE (2008) Mouse hepatitis virus infection of the CNS: a model for defense, disease, and repair". Frontiers in Bioscience 13(13): 4393-4406.
- Liu P, Shi L, Zhang W, He J, Liu C, Zhao C, et al. (2017) Prevalence and genetic diversity analysis of human coronaviruses among cross-border children. Virology Journal 14 (1): 230.
- Forgie S, Marrie TJ (2009) Healthcare-associated atypical pneumonia". Seminars in Respiratory and Critical Care Medicine 30 (1): 67-85.
- Sorush Niknamian (2020) Introducing Covid-19 as an Evolutionary Metabolic Infectious Disease (Emid) the Prime Cause and Representing Alternative Treatment for COVID-19 (SARS-CoV-2). J B Bio Engine 1(1).
- Ivanov IP, Atkins JF (2007) Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Research 35 (6): 1842-1858.
- Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, et al. (1988) Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331(6153): 280-283.
- Jacks T, Madhani HD, Masiarz FR, Varmus HE (1988) Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55(3): 447-458.
- Venkatesan M, Silver LL, Nossal NG (1982) Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J. Biol. Chem 257(20): 12426-12434.
- Matson SW, Tabor S, Richardson CC (1983) The gene 4 protein of bacteriophage T7. Characterization of helicase activity". J. Biol. Chem 258(22): 14017-14024.
- Steimer L, Klostermeier D (2012) RNA helicases in infection and disease. RNA Biology 9 (6): 751-771.
- Jankowsky E, Fairman-Williams ME (2010) An introduction to RNA helicases: superfamilies, families, and major themes". In Jankowsky E (ed.). RNA Helicases (RSC Biomolecular Sciences). Cambridge, England: Royal Society of Chemistry p. 5.
- Ranji, A, Boris-Lawrie K (2010) RNA helicases: Emerging roles in viral replication and the host innate response. RNA Biology 7 (6): 775-787.
- Jankowsky E (2011) RNA helicases at work: binding and rearranging. Trends Biochem Sci 36 (1): 19-29.
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E (2007) DEAD-box proteins unwind duplexes by local strand separation. Mol Cell 28(2): 253-263.
- Liu F, Putnam A, Jankowsky E (2009) ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA 105(51): 20209-202014.
- Jarmoskaite I, Russell R (2011) DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA 2 (1): 135-152.
- Lainé JP, Mocquet V, Egly JM (2006) TFIIH enzymatic activities in transcription and nucleotide excision repair. Meth. Enzymol. Methods in Enzymology 408: 246-263.
- Darwin Charles (1871) Pangenesis. Nature 3(78): 502-503.
- Holterhoff, Kate (2014) The History and Reception of Charles Darwin's Hypothesis of Pangenesis". Journal of the History of Biology 47(4): 661-695.
- Liu Yongsheng (2008) A new perspective on Darwin's Pangenesis. Biological Reviews 83(2): 141-149.
- Orr HA (1996) Dobzhansky, Bateson, and the genetics of speciation. Genetics 144 (4): 1331-1335.
- Bateson W (1909) Heredity and variation in modern lights Darwin and Modern Science 85-101.
- Dobzhansky T (1934) Studies on Hybrid Sterility. I. Spermatogenesis in pure and hybrid Drosophila pseudoobscura. Zeitschrift für Zellforschung und mikroskopische Anatomie 21(2): 169-221.
- Coyne, Barton, Turelli (1997) Perspective: a critique of Sewall Wright's shifting balance theory of evolution. Evolution 51(3): 643-671.
- Coyne, Barton, Turelli (2000) Is Wright's shifting balance process important in evolution?". Evolution 54(1): 306-317.
- Gould SJ (1980) GG Simpson, Paleontology and the Modern Synthesis." In E. Mayr and W. B. Provine, eds., The Evolutionary Synthesis. Cambridge MA: Harvard University Press, pp. 153-172.
- Simpson GG (1954) The Major Features of Evolution. New York: Columbia Univ. Press, 35(4): 600-601.
- Simpson GG (1944) Tempo and Mode in Evolution. New York: Columbia Univ. Press 3(2): 208-209.
- Grant, Verne (1963) The Origin of Adaptations. New York and London: Columbia University Press. 606.
- Lewis Harlan, Raven Peter H (1958) Rapid Evolution in Clarkia. Evolution 12(3): 319-336.
- Lewis Harlan (1962) Catastrophic Selection as a Factor in Speciation. Evolution 16(3): 257-271.
- Gottlieb LD (2003) Rethinking classic examples of recent speciation in plants. New Phytologist 161(1): 71-82.
- Gottlieb LD (1973) Genetic Differentiation, Sympatric Speciation, and the Origin of a Diploid Species of Stephanomeria. American Journal of Botany 60(6): 545-553.
- Simpson GG (1953) The Major Features of Evolution p. 390.
- Gould SJ (1983) The hardening of the Modern Synthesis In: Marjorie Grene (eds.) Dimensions of Darwinism. Cambridge UK Cambridge University Press pp. 71-93.
- Marra MA, Jones SJ, Astell CR, Robert A Holt, Angela Brooks-Wilson, et al. (2003) The genome sequence of the SARS-associated coronavirus. Science 300: 1399-1404.
- Zhong NS, Zeng GQ (2003) Our strategies for fighting severe acute respiratory syndrome (SARS). Am J Respir Crit Care Med 168: 7-9.
- Bell D, Jenkins P, Hall J (2003) World Health Organization global conference on the severe acute respiratory syndrome. Emerg Infect Dis 9(9): 1191-1192.
- Aleanizy FS, Mohmed N, Alqahtani FY, El Hadi Mohamed RA (2017) An outbreak of Middle East respiratory syndrome coronavirus in Saudi Arabia: a retrospective study. BMC Infect Dis pp. 17: 23.
- Haagmans BL, Al Dhahiry SH, Reusken CB, V Stalin Raj, Monica Galiano, et al. (2014) Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis 14: 140-145.
- Conforti C, Giuffrida R, Dianzani C, Di Meo N, Zalaudek I, et al. (2021) COVID-19, and psoriasis: is it time to limit treatment with immunosuppressants? A call for action. Dermatol Ther 11: e13298.
- Yan Y, Chen H, Chen L, Bo Cheng, Ping Diao, et al. (2020) Consensus of Chinese experts on protection of skin and mucous membrane barrier for healthcare workers fight-ing against coronavirus disease 2019. Dermatol Ther 13: e13310.
- Li Q, Guan X, Wu P (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382: 119 9-1207.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...