Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-5945
Research Article(ISSN: 2638-5945) 
Implication of Integrated in Silico Docking and Moma Simulation for the Development of Curcuminoids as Sphingosine Kinase 1 (SK1) Inhibitors for Cancer Treatment Volume 3 - Issue 3
Noboru Motohashi1, Anuradha Vanam2, Jyothirmayi Vadapalli3 and Rao Gollapudi4*
- 1Meiji Pharmaceutical University, Japan
- 2Sri Venkateswara University, India
- 3Acharya Nagarjuna University, India
- 4University of Kansas, USA
Received:January 02, 2020 Published: January 23, 2020
Corresponding author: Rao Gollapudi, University of Kansas, USA
DOI: 10.32474/OAJOM.2020.03.000165
Abstract
Sphingosine kinase 1 (SphK1) enzyme catalyzes the phosphorylation of sphingosine into sphingosine-1-phosphate (S1P). S1P plays an important role in diverse biological processes. In numerous human cancers, overexpression of SphK1 led to the severity of disease, drug resistance and reduced patient survival. Furthermore, SphK1 increased activity was connected to play a crucial role in immunological responses such as asthma, rheumatoid arthritis and sepsis. Inhibitors of SphK1 enzyme helps in reducing the adverse effects caused by over expression of SpkH1. Hence, there is a growing demand for the discovery of new SphK1 inhibitors. Curcumin (5) a major constituent of turmeric rhizomes, displayed the inhibition activity of cancers by regulating multiple cellular signaling pathways. Henceforth, the present molecular docking study suggested that curcumin (5) could be valuable SphK1 inhibitor as a result of its binding affinity to the SphK1 active site similar to that of CTK8F1052 (4). Further exploration of curcumin (5) efficacy in inhibiting the SphK1 activity is necessary to support computational docking findings.
Keywords: Sphingosine Kinase1; Sphingosine-1-Phosphate; Adenosine Triphosphate; CTK8F1052; Curcumin; Cyclophosphamide; Docetaxel; Doxorubicin; Cellular Pathways; Cancer; Lipinski’s Rule of Five; In Silico Docking Studies.
List of Abbreviations: SIP: Sphingosine-1-Phosphate; GTP-G: Guanosine Triphosphate; G-Protein: Binding Protein; GRK: Coupled Receptor Kinases; S1P: Sphingosine-1-Phosphate; SphK: Sphingosine Kinase; ATP: Adenosine Triphosphate; TNBC: Triple Negative Breast Cancer; CVDs: Cardiovascular Disorders; SK: Sphingosine Kinase; ER: Estrogen Receptor; PDB: Protein Data Bank; MVD: Molegro Virtual Docking; MMFF: Merck Molecular Force Field
Introduction
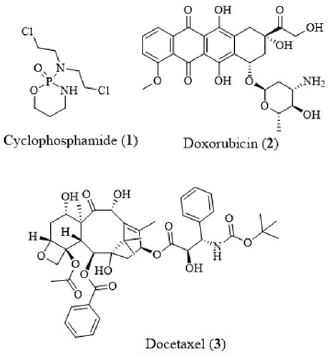
Sphingosine kinase 1 (SphK1) is generally locolaized in cytosol and belong to a class of guanosine triphosphate (GTP. G)-binding protein (G-protein)-coupled receptor kinases (GRK). In humans, SphK1 is encoded by SPHK1 gene located in chromosome 17. SphK1 catalyzed the phosphorylation of sphingosine into sphingosine- 1-phosphate (S1P). S1P was recognized as an important player in multiple biological developments, including cancer and inflammation [1-5]. Sphingosine kinase (SphK) catalyzed adenosine triphosphate (ATP)-dependent phosphorylation of sphingosine lipid followed by alteration of the last step in production of S1P. SphK subsists in two isoforms; SphK1 and SphK2. Different sphingolipid metabolites operated as supplementary lipid messengers which participated in miscellaneous cellular processes, comprising of cellular migration, proliferation, and apoptosis. SphKs were recognized as biomarkers in different types of cancer by promoting angiogenesis and tumorigenesis [6]. Amplified expression of sphingosine kinase 1 (SphK) was concomitant with breast cancer progression including the resistance to drug therapies [7-9]. Furthermore, exogenous administration of S1P amplified the cytotoxic potential of chemotherapy drugs, cyclophosphamide (1), doxorubicin (2) and docetaxel (3) against breast cancer metastatic cell lines (Figure 1) [10]. Nevertheless, SphK1 was up-regulated in numerous human cancers where there was a correlation between SphK1 expression, severity of disease, drug resistance and reduced patient survival [11-13]. SphK1 activity was connected to play a crucial role in immunological responses such as asthma, rheumatoid arthritis and sepsis [14-16]. Subsequently, the application of SphK1 antagonists inhibited cell growth and induced apoptosis in different human cancer cell lines. In addition, SK1 antagonists exhibited radio-sensitizing effects on triple-negative breast cancer (TNBC) cell lines, increasing the anti-proliferative and pro-apoptotic effects that were induced by ionizing radiation [17]. SphK1 inhibition also stimulated apoptosis and reduced cell proliferation in both in vivo and in vitro TNBC models [18]. Lately, SphK1 was recognized as a critical player in the development of cancer, inflammation, autoimmune, neurological and cardiovascular disorders (CVDs). Over expression of SphK1 was related to cancer prognosis and development. The SphK1 inhibitor, CTK8F1052 (SHI-II) (4) suppressed cell growth and induced apoptosis in human hepatoma HepG2 cells. CTK8F1052 (4)-initiated down-regulation of HepG2 cell proliferation was associated with Wnt signaling pathway which is network of proteins involved in embryonic development and cancer through Wnt5A-mediated β-catenin degradation [19]. Therefore, there is a prodigious interest in the development of new SphK1 inhibitors which could serve as potential players in the cancer treatment.
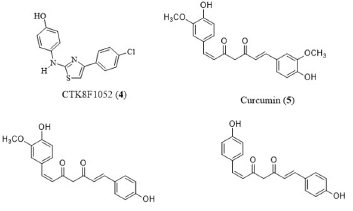
Targeting sphingosine-1-phosphate production decrease with inhibition drugs is a practical approach to retune the cell signaling aberrations that resulted in endocrine resistance. This approach could contribute to increased efficacy of current therapeutic practices by re-sensitizing cells to endocrine therapy (hormone therapy). Crosstalk between sphingolipid signaling pathways and steroid hormones is a feasible therapeutic target. In case of solid tumor cancers, sphingosine kinase (SK) is a key enzyme metabolizing pro-apoptotic ceramide to pro-survival sphingosine-1-phosphate (S1P) which is a likely therapeutic target. Sphk1/2 selective inhibitor, CTK8F1052 (4) impeded breast cancer viability, clonogenic survival and proliferation and acted as novel estrogen receptor (ER) signaling inhibitors in breast carcinoma. Curcumin (curcumin-I. 5), demethoxycurcumin (curcumin-II) (6) and bisdemethoxycurcumin (curcumin-III) (7) are the major phytochemicals present in turmeric rhizomes that are responsible for wide range of health benefits including antiinflammatory, antioxidant and antitumor activities (Figure 2). Curcumin (5) inhibited animal and human cancers by regulating multiple cellular signaling pathways [20-22]. Recently, turmeric and curcuminoids were engaged in clinical trials for the evaluation of their effectiveness in the treatment of multiple diseases. In addition, curcumin displayed synergism with cytotoxic drugs in the treatment of different cancers [23-25]. Henceforth, in silico approach was selected to evaluate the effectiveness of curcuminoids in inhibiting SphK1 thereby preventing the phosphorylation of sphingosine which was identified as one of the principles causes of chemo resistance to selected chemotherapeutic drugs [5-33].
Figure 2: Structures of CTK8F1052 (4), curcumin (curcumin-I) (5), demethoxycurcumin (curcumin-II) (6) and bisdemethoxycurcumin (curcumin-III) (7).
Materials and Methods
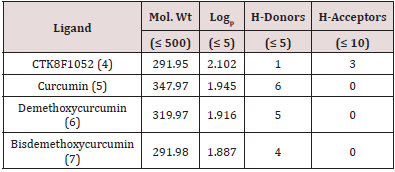
Lipinski’s rule of five in the initial stages of drug discovery, the drug-likeness of lead molecules is essential to reduce the experimental cost. The best lead candidate should not deviate more than one criteria as defined by the “Lipinski’s rule of five” (RO5). RO5 includes molecular weight (no greater than 500 Daltons or g/ mol), octanol-water partition coefficient (Po/w) (Logp of 5 or less), hydrogen bond donors (5 or less) and acceptors (10 or less). The drug likeliness tool (DruLiTo) software was used to determine the physicochemical characteristics of CTK8F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7) (Table 1). All four compounds qualified majority of Lipinski criteria (Table1) [34]. Docking Between Sphk1 Enzyme and CTK8F1052 (4), Curcumin (5), Demethoxycurcumin (6) And Bisdemethoxycurcumin (7). The ligand/protein simulated interactions of CTK8F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7) with SphK1 enzyme were determined by employing AutoDock Vina (Molecular Graphics Lab, La Jolla, CA, USA) [35]. The AutoDock Vina software selected for organizing the target protein in a rigid conformation while the ligands were allowed to be malleable to the target. The lowest binding affinity of the ligand docked to the receptor was determined by software through different confirmations of each ligand. The subsequent lowest binding energy of the docking posture for each ligand was selected for further evaluation.
Table 1: Review of the literature – Research Articles of metastatic bone or soft tissue sarcoma to the small bowel.
Protein Preparation
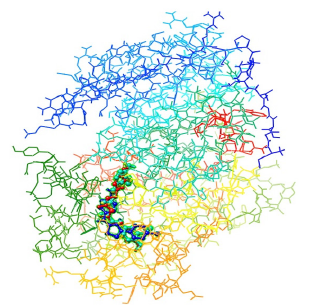
For this docking study, the three-dimensional (3D) crystal structure of SphK1 and CTK8F1052 (4) complex (PDBID: 3VZC) was retrieved from Protein Data Bank (PDB) and chain-A was selected for the docking study. The cofactor CTK8F1052 (4) (Chain A) and water molecules were removed from the 3VZC chain A PBD file using Discovery Studio 4.5 (Dassault Systemes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego, USA). A potential binding cavity selected for this docking study was identified on SphK1 receptor (3VZC chain A) with a volume of 631.808 Ao using Molegro Virtual Docking 2011. 4.3.0 (MVD) software (Figure 3). Subsequently, polar hydrogen atoms were added to the protein model using AutoDock tools 4.2.6 and PyRx v0.8 software. The docking grid was selected using PyRx interlinked with AutoDock Vina [35,36].
Figure 3: The PDB structure of SphK1 (3VZC chain A) with active binding cavity was shown in coloured in opal blue.
Docking between SphK1 Enzyme and CTK8F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7)
Docking simulations were performed by defining the grid (Box size (X,Y,Z): 28.2547 × 40.4360 × 31.7637 Å and box center: center x = 0.7848, center y = -32.5874, center z = -5.5194, respectively) with exhaustiveness value of 8 covering the ligand binding site. PyMol v1.3 (Schrodinger, New York, NY, USA), Chimera (UCSF, San Francisco, CA, USA) and Discovery Studio software were used for visual inspection and graphical illustrations of the docking results. The pdb structure of SphK1 complex with CTK8F1052 reported earlier was downloaded (3VZC) from Protein Data Bank and the prepared SphK1 chain A were shown in (Figure 4) [37].
Preparation of Ligands
The structures of CTK6F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7), were drawn using ChemDraw Professional 18.2 (CambridgeSoft) followed by the conversion to PDB format using Chem3D (CambridgeSoft). Avogadro software was used for molecular geometry optimization and Merck molecular force field (MMFF) of ligands. Those ligands were saved in Protein Data Bank (PDB) file format for molecular docking studies. An open-source molecular builder and visualization tool.
SphK1 and Ligands Docking
PyRx virtual screening software interlinked with AutoDock Vina was used for SphK1dockings with CTK6F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7). A set of possible confirmations of each ligand was generated by docking with different orientations of ligands within the SphK1 binding site. The protein was kept rigid while the ligands were left flexible during the docking process. Upon the completion of docking process, the ligand confirmations with highest binding affinity and lowest docked energies to the target were selected. The hydrogen bonds, bond lengths and hydrophobic interactions between protein (SpkH1) and ligands CTK6F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7) were determined by employing LigPlot and Discovery Studio software. The docking complexes of CTK8F1052 and curcumin with SphK1 (3VZC chain A) receptor were subjected for further evaluation by performing unbinding simulations on MoMA LigPath server and the results of these complexes received from the server were compared and evaluated.
Unbinding Simulations of Target / Ligand Complexes
Protein-ligand unbinding simulation: MoMA-LigPath is a web server which computes Molecular Motion Algorithms (MoMA). The server simulates the ligand unbinding from the binding site of the target to the surface of the target. In addition, the server studies the flexibility of protein side-chains, ligands and includes only statistical limitations. This process generates mechanistic data on the ligand pathway to the binding site from the surface of the protein and/or from the binding site to the surface. The Program offers molecular interaction graphics, leading the ligand from the surface of the protein to the binding site. In the process, it identifies certain residues that play a vital role in ligand binding or in driving the ligand to the binding site, in spite of being away from the binding site. The docked molecular complexes of CTK8F1052 and curcumin of SphK1 rcceptor were selected for unbinding simulations by using MoMA LigPath [38-40].
Results and Discussion
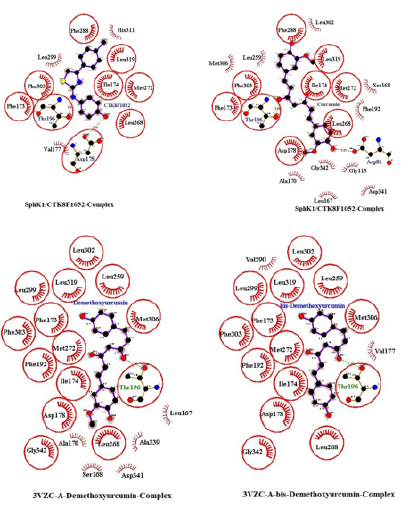
Previous in silico docking studies disclosed curcumin (5) inhibitory affects by binding to proteins, enzymes and receptors targets (EGF2, GST-PI, PDGFA, MMP-3, HER-2, ERBB2, C-CRC, FTO) which were involved in regulating cancer, inflammation and obesity through cellular signaling pathways [41-44]. The interpretation of protein-ligand interactions plays an important role in structural based drug discovery. TheSphK1 inhibitor, CTK8F1052 (4) was docked on SphK1 receptor with binding energy value of -9 kcal/mol. Curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7) were docked on the same SpkH1 receptor with binding energies of -9.8, -9.7 and -9.5 kcal/mol. Curcumin (5) interactions with SphK1 were similar to that of CTK8F1052 (4) binding patterns with SphK1 kinase as described in the SphK1 crystallographic data [37]. The enclosed lipid pocket of SpkH1 was subjugated by curcumin (5) where the alkyl chain of sphingosine was bound to the phenol ring situated at the end of the pocket. The phenol moiety of curcumin (5) pointed toward the opening in the cleft between N-terminal domain (residues 9-150 and 357-364, containing the C1-C3 domains), adopted the a/b fold which comprised six a helices (a1-a6) and six b strands (b1-b5 and b17). The entire formation resembled the dinucleotide binding motif of a Rossmann fold of a three-layer a/b/a sandwich and included the C-terminal domain (residues 151-356, containing the C4 and C5 domains) which comprised 11 b strands and four helices as described between curcumin (5) and SpkH1 interactions. The hydroxyl moiety of the phenol ring was involved in wander Waal attraction to Asp178 of helix a7, leading to a closed conformation for the lipid gate. Furthermore, the C-5 ketone group of curcumin (5) was involved in hydrogen bond with the hydroxyl of Thr196 of helix a8. Interestingly, C-4 hydroxyl group of curcumin (5) formed hydrogen binding with ASP 81 of N-terminal domain preventing the interaction of sphingosine where the C (2) amine of sphingosine was required to be located nearly with equal distance between aspartates 81 and 178. Curcumin (5) bonded with SphK1 in a hydrophobic pocket that was formed between the strands b10- b12 of the b-sandwich in the C-terminal domain and helices a7- a9 where and the hydrophobic tail of sphingosine interacted with SpkH1.
The rest of the molecule was involved in van der Waals interactions with a similar set of nonpolar residues that recognized the hydrophobic tail of the lipid substrate of SpkH1 similar to CTK8F1052 (4) with additional interactions (Figure 4). Consequently, curcumin (5) could be a lipid substrate competitive inhibitor of SpkH1 preventing the interactions of sphingosine with SpkH1 thereby preventing sphingosine phosphorylation. Demethoxycurcumin (6) and bisdemethoxycurcumin (7) interactions with SpkH1 receptor were similar to that of curcumin (5) except the missing of hydrogen bonds between the two curcumonoids with ASP81 of SpkH1 suggesting that curcumin (5) could be a better inhibitor of SpkH1 compared to other two curcuminoids. The interactions of CTK8F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7) to the receptor protein, SpkH1 are displayed in (Figure 5).
Figure 4: A: The PDB structure of SphK1 (3VZC) with labelled as a, b, c, d, e and f chains; B: The PDB structure of prepared Sphk (3VZC chain A) for docking study.
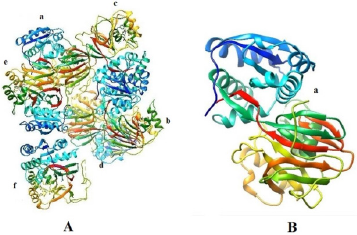
Figure 5: PROTEIN LIGAND COMPLEXES OF: TOP LEFT CTK8F1052 /Sphk1 compiles; Top RIGHT: Curcumin/ Sphk1 Complex; Bottom Left: Demethoxy Curcumin and Bottom Right: Bis- demethoxy Curcumin
Unbinding Simulation of Sphk1 Receptor/CTK8F1052 Complex
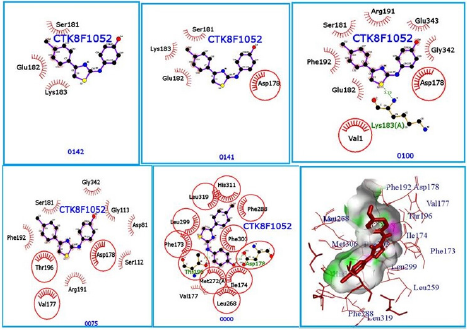
The unbinding simulation phases of CTK8F1052 (4) with increasing number of molecular interactions while progressively approaching towards the binding site of SphK1 are shown in (Figure 6) (A-1 to A-5). The docking phase display the molecular interactions of SphK1 with CTK8F1052 (4) bound on theexterior region of SphK1 cavity by hydrophobic interactions. In addition, CTK8F1052 (4) was involved in interaction through Asp 178. The amino acid, Asp 178 was common in all phases of unbinding simulation of CTK8F1052 (4) which emphasized the importance of Asp 178 in stabilizing SphK1/ CTK8F1052 (4) complex. The whole protein is displayed in cartoon representation (A-6). The interacting ligand and receptor residues are labelled and shown as surface in different colours (Figure 6).
Figure 6: CTK8F1052 (1) binding to SphK1. Panels A-1 to A-5 show the unbinding simulation phases of CTK8F1052: A-5 is farthest from the binding site, A-1 is the closest to the binding site, and A-6 is the binding site phase. The hydrogen bonds are shown as green-dashed lines with indicated bond length and the residues involved in hydrophobic interactions are shown as red arcs. A-6: The SphK1 receptor is displayed in cartoon representation, CTK8F1052 (1) in sticks and coloured in red. The interacting residues are labelled and shown as surface in different colours.
Unbinding Simulation of Sphk1 Receptor/Curcumin Complex
The unbinding simulation phases of curcumin (5) with increasing number of molecular interactions while progressively approaching towards the binding site of SphK1 are shown in (Figure 7) (A-1 to A-5). The docking phase display the molecular interactions of SphK1with curcumin (5) bound on the exterior region of SphK1 cavity by hydrophobic interactions. In addition, curcumin (5) was involved in interaction through Asp 178. The amino acid, Asp 178 was common in all phases of unbinding simulation of curcumin (5) which emphasized the importance of Asp 178 in stabilizing SphK1/ curcumin (5) complex. The whole protein is displayed in cartoon representation (A-6). The interacting ligand and receptor residues are labelled and shown as surface in different colors (Figure 7). Collectively, the computational results from in silico docking of curcumin (5) and CTK8F1052 on SphK1 receptor suggested that curcumin (5) could be a potential SphK1 inhibitor with binding affinity comparable to CTK8F1052. Furthermore, these findings were verified through unbinding simulations of docked ligands/receptor complexes on MoMA-LigPath (a web server). Consequently, curcumin (5) was selected for further investigation to evaluate its effectiveness as a competent inhibitor of SphK1. A graphical representation of docked CTK8F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7) on SphK1 receptor is shown in (Figure 8).
Figure 7: CTK8F1052 (1) binding to SphK1. Panels A-1 to A-5 show the unbinding simulation phases of CTK8F1052: A-1 is farthest from the binding site, A-5 is the closest to the binding site. The hydrogen bonds are shown as green-dashed lines with indicated bond length and the residues involved in hydrophobic interactions are shown as red arcs. A-6: The SphK1 receptor is displayed in cartoon representation, CTK8F1052 (1) in sticks and coloured in red. The interacting residues are labelled and shown as surface in different colours.
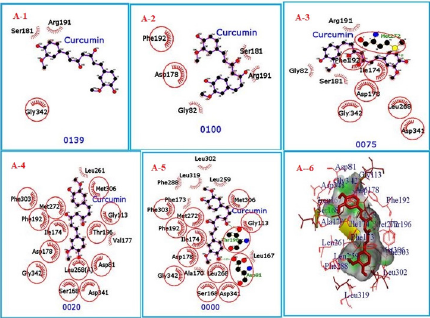
Figure 8: Interactions of CTK8F1052 (4) (royral blue), curcumin (5) (turquoise blue), demethoxycurcumin (6) (yellow) and bisdemethoxycurcumin (7) (red) with SphK1.
Conclusion
SphK1 is a lipid kinase that catalyzes the conversion of sphingosine to sphingosine-1-phosphate (S1P). SphK1 performs a key role in lymphocyte trafficking, angiogenesis, and response to apoptotic stimuli. Furthermore, SphK1 is essential enzyme in altering the S1P levels in cells and hence transpires as an important regulator for multiple cellular functions making it a potential target in the drug discovery process. Our present in silico docking study suggested that CTK8F1052 (4), curcumin (5), demethoxycurcumin (6) and bisdemethoxycurcumin (7) displayed interactions with SphK1 through hydrogen bonds and van der Waals with binding energies (Kcal) of -9, -9.8, 9.7 and -9.5, respectively. The binding interactions between curcumin (5) and SphK1 are similar to that of CTK8F1052 (4) interactions with SphK1 for the inhibition of SphK1. Moreover, curcumin (5) displayed no toxicity up to 8 g per day in the treatment of multiple diseases [45]. These findings could be helpful in the development of targeted therapeutic strategy for cancer treatment. Henceforth, curcumin (5) could be a potential SpkH1 inhibitor as CTK8F1052 (4).
References
- Beaven MA (2007) Division of labor: specialization of sphingosine kinases in mast cells. Immunity 26(3): 271-273.
- Brinkmann V, Billich A, Baumruker T, Heining P, Schmouder R, et al. (2010) Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat Rev Drug Discov 9(11): 883-897.
- Pyne NJ, Pyne S (2010) Sphingosine 1-phosphate and cancer. Nat Rev Cancer 10: 489-503.
- Maceyka M, Harikumar KB, Milstien S, Spiegel S (2012) Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol 22(1): 50-60.
- Pyne NJ, Tonelli F, Lim KG, Long J, Edwards J et al. (2012) Targeting sphingosine kinase 1 in cancer. Adv Biol Regul 52(1): 31-38.
- Olivera A, Spiegel S (1993) Sphingosine-1-phosphate as second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature 365(6446): 557-560.
- Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, et al. (1996) Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 381(6585): 800-803.
- English D, Kovala AT, Welch Z, Harvey KA, Siddiqui RA, et al. (1999) Induction of endothelial cell chemotaxis by sphingosine 1-phosphate and stabilization of endothelial monolayer barrier function by lysophosphatidic acid, potential mediators of hematopoietic angiogenesis. J Hematother Stem Cell Res 8(6): 627-634.
- Rosen H and Goetzl EJ (2005) Sphingosine 1-phosphate and its receptors: an autocrine and paracrine network. Nat Rev Immune 5(7): 560-570.
- Kihara A, Mitsutake S, Mizutani, Igarashi Y (2007) Metabolism and biological functions of two phosphorylated sphingolipids, sphingosine 1-phosphate and ceramide 1-phosphate. Prog Lipid Res 46(2): 126-144.
- Hannun YA, Obeid LM (2008) Principles of bioactive lipid signalling: lessons from sphingolpids. Nat Rev Mol Cell Biol 9(2): 139-150.
- Chi H (2011) Sphingosine-1-phosphate and immune regulation: trafficking and beyond. Trends Pharmacol Sci 32(1): 16-24.
- Spiegel S and Milstien S (2011) The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol 11(6): 403-415.
- Maceyka M, Payne SG, Milstien S, Spiegel S (2002) Sphingosine kinase, sphingosine-1-phosphate and apoptosis. Biochim Biophys Acta 1585(2-3): 193-201.
- Antoon JW, White MD, Driver JL, Burow ME, Beckman BS, et al. (2012) Sphingosine kinase isoforms as a therapeutic target in endocrine therapy resistant luminal and basal-A breast cancer. Exp Biol Med 237(7): 832-844.
- Yagoub D, Wilkins MR, Lay AJ, Kaczorowski DC, Hatoum D, et al. (2014) Sphingosine kinase 1 isoform-specific interactions in breast cancer. Mol Endocrinol 28(11): 1899-1915.
- Tsuchida J, Nagahashi M, Takabe K, Wakai T (2017) Clinical impact of sphingosine-1-phosphate in breast cancer. Mediat Inflamm pp. 2076239.
- Sultan A, Ling B, Zhang H, Ma B, Michel D, et al. (2013) Synergistic effect between sphingosine-1-phosphate and chemotherapy drugs against human brain-metastasized breast cancer MDA-MB-361 cells. J Cancer 4(4): 315-319.
- Baker DA, Barth J, Chang R, Obeid LM, Gilkeson GS, et al. (2010) Genetic sphingosine kinase 1 deficiency significantly decreases synovial inflammation and joint erosons in murine TNF- alpha induced arthritis. J Immunol 185(4): 2570-2579.
- Puneet P, Yap CT, Wong L, Lam Y, Koh, et al. (2010) SphK1 regulate proinflammatory responses associated with endotoxin and polymicrobial sepsis. Science 328(5983): 1290-1294.
- Price MM, Oskeritzian CA, Falanga YT, Harikumar KB, Allegood JC, et al. (2013) A specific sphingosine kinase 1 inhibitor attenuates airway hyperresponsiveness and inflammation in mast cell-dependent murinemodel of allergic asthma. Allergy Clin Imunol 131(2): 501-511.
- Marvaso G, Barone A, Amodio N, Raimondi L, Agosti V, et al. (2014) Sphingosine analog fingolimod (fty720) increases radiation sensitivity of human breast cancer cells in vitro. Cancer Biol Ther 15(6): 797-805.
- Liu H, Zhang CX, Ma Y, He HW, Wang JP, et al. (2016) SphK1 inhibitor SKI II inhibits the proliferation of human hepatome HepG2 cells via the Wnt5A/B-catenin signaling pathway. Life Sci 151: 23-29.
- Barati N, Momtazi Borojeni AA, Majeed M, Sahebkar A (2019) Potential therapeutic effects of curcumin in gastric cancer. J Cell Physiol 234(3): 2317-2328.
- Jalili Nik M, Soltani A, Moussavi S, Ghayour Mobarhan M, Ferns GA, et al. (2018) Current status and future prospective of Curcumin as a potential therapeutic agent in the treatment of colorectal cancer. J Cellular Physiol 233(9): 6337-6345.
- Kunnumakkara AB, Anand P, Aggarwal BB (2008) Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett 269(2): 199-225.
- He B, Wei W, Liu J, Xu Y, Xhao G, et al. (2017) Synergistic anticancer effects of curcumin and chemotherapy regimen FP in human gastric cancer MGC-803 cells. Oncol Lettt PP. 3387-3394.
- Rao J, Xu DR, Zheng FM, Long ZJ, Huang S, et al. (2011) Curcumin reduces expression of Bcl-2 leading to apoptosis in daunorubusin insensitive CD34+ acute myeloid leukemia cell lines and primary sorted CD34+ acute meloid leukemis cells. J Transl Med 9: 71.
- Alemi A, Zavar Reza J, Haghiralsadat F, Zarei Jaliani H, Haghi Karamallah M, et al. (2018) Paclitaxel and curcumin coadministration of novel cationic PEGylated niosomal formulations exhibited enhanced synergistic antitumor efficacy. J. Nanobiotechnology 16(1): 28.
- Ponnusamy S, Meyers-Needham M, Senkal CE, Saddoughi SA, Sentelle D, et al. (2010) Sphingolipids and cancer: ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol 6(10): 1603-1624.
- Gault CR, Obeid LM (2011) Still benched on its way to the bedside: sphingosine kinase 1 as an emerging target in cancer chemotherapy. Crit Rev Biochem Mol Biol 46(4): 342-351.
- Pitman MR, Pitson SM (2010) Inhibitors of the sphingosine kinase pathway as potential therapeutics. Curr Cancer Drug Targets 10(4): 354-367.
- Shida D, Takabe K, Kapitonov D, Milstien S, Spiegel S, et al. (2008) Targeting SphK1 as a new strategy against cancer. Curr Drug Targets 9(8): 662-673.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1-3): 3-26.
- Trott O, Olson AJ (2010) Auto Dock vina: improving the speed and accuracy of docking with a new drug scoring function. J Comput Chem 31(2): 455-461.
- Dallakyan S, Oleson A (2015) Small molecule library screening by docking with PyRx. Methods Mol Biol 1263: 243-250.
- Wang Z, Min X, Xiao SH, Johnstone S, Romanow W, et al. (2013) Molecular basis of sphingosine kinase 1 substrate recognition and catalysis. Structure 21(5): 798-809.
- Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, et al. (2012) Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4: 17.
- Cortes J, Le DT, Iehl R, Simeon T (2010) Simulating ligand-induced conformational changes in proteins using a mechanical disassembly method. Physical Chem Chem Phys 12(29): 8268-8276.
- Devaurs D, Bouard L, Vaisset M, Zanon C, Al Bluwi I, et al. (2013) MoMA LigPath: a web server to simulate protein ligand unbinding. Nucleic Acids Research 41(W1): W297-W302.
- Jerah A, Hobani Y, Kumar BV, Bidwai A (2015) Curcumin binds in silico to anti-cancer drug target enzyme MMP-3 (human steomelysin-1) with affinity comparable to two known inhibitors of the enzyme. Bioinformation 11(8): 387-392.
- Maharajanakatti AB, Murthy G, Sharma N, Skariyachan S (2014) Exploring inhibitory potential of curcumin against various cancer targets by in silico virtual screening. Interdiscip Sci 6(1): 13-24.
- Pusphalatha P, Selvamuthukumar S, Kilimozhi C (2017) Comparative in silico docking analysis of curcumin and resveratrol on breast cancer proteins and their synergistic effect on MCF-7 cell line. J Young Pharm 9(4): 480-485.
- Archana P, Sathish Kumar N, Bharathi N (2010) In silico docking analysis of curcumin-an inhibitor for obesity. Int J Pharm Bio Sci 1(4): 224-235.
- Cheng AL, Hsu CH, Lin JK, Hsu MM, Ho YF, et al. (2001) Phase 1 clinical trail of curcumin, a chemopreventive agent in patients with high-risk or pre-malignant lesions. Anticancer Res 21(4B): 2895-2900.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...