Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-5945
Case Report(ISSN: 2638-5945) 
Carbapenem-Resistant Bacteria in Oncology Clinic Volume 2 - Issue 3
Victoria Aginova* and Natalia Dmitrieva
- Department of Oncology, College of Medicine, Moscow, Russia
Received: January 22, 2019 Published: February 04, 2019
Corresponding author: Victoria Aginova, Department of Oncology, College of Medicine, Moscow, Russia
DOI: 10.32474/OAJOM.2019.02.000146
Abstract
The present study was objected to check the awareness about SARS among scholars of university. The pupil of Bahauddin Zakariya university Multan acted as the subjects for this research. Total of 109 subjects showed consent to be the part of this study. Severe acute respiratory syndrome (SARS) is considered a deadly animal virus which, perhaps originated from bats and then it was spread to other animals. According to a report, the humans were affected by SARS very firstly in 2002 in Guangdong, china. a report by World health organization states that 8098 people were affected by SARS in an outbreak during 2003, out of which 774 died. It was first observed as an influenza like disease with headache, fever leading to failure of respiratory system and sometimes death of the patient. The early symptoms of SARS are headache, body ache, high fever, coughing and sneezing, feeling difficulty in breathing leading to asthma sometimes. Treatment of SARS includes taking antibiotics (to prevent pneumonia) and some antiviral medicines. Present research helped in knowing about the knowledge of SARS in university students. This study helped in estimating that 84.40% pupil consider SARS as a viral disease, 12.84% students think it’s a genetic problem while 9.17% suppose it’s a metabolic error.
Keywords: SARS (severe acute respiratory syndrome); Pupil; Ailment; Viral; Metabolic; Pandemics; Steroids
Introduction
The occurrence of carbapenem-resistant (CarR) bacteria is a serious problem for the treatment of potentially fatal infections. According to estimates of most authors the mortality rate for infections caused by the CarR microorganisms is 24%-70% [1-4]. They are a growing concern in global public-health due to their associative resistance to all or almost all β-lactams and other classes of antibiotics, such as aminoglycosides, fluoroquinolones and cotrimoxazole. Treatment options are limited and there is still no consensus on optimal treatment regimens [5-7]. Therefore, regular monitoring of carbapenem-resistant strains of microorganisms and followed by the determination of the molecular mechanism of resistance is extremely important to prevent the spread of such bacteria and to select the optimal antibiotic therapy in any clinics. The purpose of this study was to determine the species composition of the carbapenem-resistant strains of microorganisms isolated from the biological materials of patients at an oncology clinic, to identify strains producing carbapenemases and to determine the types of carbapenemases.
Materials and Methods
Studies were conducted in the period 2017-2018. Sources of bacterial isolates were biological materials from patients of the oncological clinic. The study took strains of gram-negative bacteria resistant (R) or intermediate resistant (I) to carbapenems. Microorganisms were identified by the method of matrixassociated laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF) and software MALDI Biotyper v.3.0 (Bruker Daltonics, Germany). Genes encoding carbapenemases of the VIM, OXA-48, OXA-40, OXA-23, NDM groups were detected by real-time polymerase chain reaction using the AmpliSense® MDR MBL-FL and AmpliSense® MDR KPC/OXA-commercial kits 48-FL” (developed by FBUN Central Research Institute of Epidemiology, Russia) and the Rotor-Gene 6000 system (Corbett Research, Australia). The strains of E. coli, K. pneumoniae, P. aeruginosa, A. baumannii from the RIACh collection (n= 14), producing known carbapenemases of the listed types, were used as positive controls. P.aeruginosa isolates tested for MBL of the VIM, IMP, NDM and Class A carbapenemases groups (GES-5-like). The strains of A.baumannii were tested for the above-mentioned metal beta-lactamase and class A carbapenemases, as well as the presence of OXA-carbapenemases of class D typical of this type (groups OXA-23, -24/40,-51, -58). Bacteria of the Enterobacteriaceae family were examined for the presence of MBL and OXA-carbapenemases (group OXA-48).
Results
The Gram-negative bacteria resistant to carbapenems were studied in the laboratory of microbiological diagnostics and treatment of infections in oncology N.N. Blokhin National Medical Research Cancer Center, Russia in conjunction with the Microbiology Laboratory of the Research Institute of Antimicrobial Chemotherapy, Russia, in 2017-2018. A total of 123 strains of the most frequently isolated microorganisms were studied (A. baumannii-35 (28.5%), P. aeruginosa-26 (21.1%), E. coli-11 (8.9%), K.рneumoniaе-37 (30.1%)). Also, the study took strains of such microorganisms as: Enterobacter cloacae, Serratia marcescens, Enterobacter asburiae, Burkholderia cepacia the relative amount of which was 10.6%. All test isolates were obtained from biological materials of oncological clinic patients. The ratio of carbapenemresistant microorganisms from different biological materials of cancer patients is presented in Figure 1.
Figure 1: The ratio of carbapenem-resistant microorganisms from various biological materials of cancer patients.
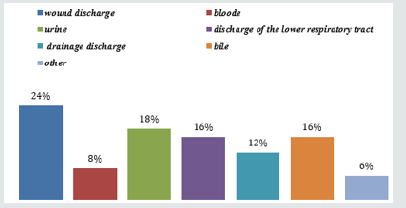
Table 1: Types of carbapenemases CarR strains of microorganisms circulating in the oncological clinic.
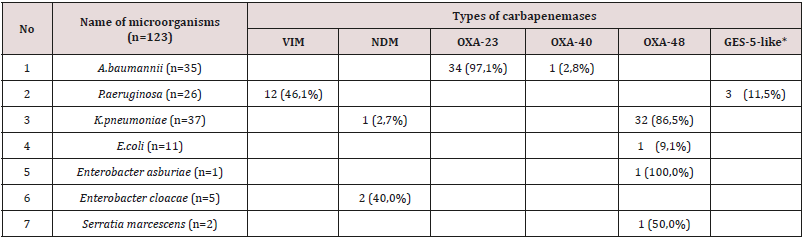
* VIM, NDM, GES, OXA-48, OXA-40, OXA-23 - name types of carbapenemases
The most frequently carbapenem-resistant strains of microorganisms were isolated from wound discharge (24%), from urine (18%), in equal amounts from the discharge of the lower respiratory tract and from bile (16%). In the discharge drainage and in the blood, CarR strains were found in 12% and 8% respectively. When conducting research, the types of carbapenemases produced by microorganism strains isolated from biological materials of cancer patients were determined. Types of carbapenemases CarR strains of microorganisms circulating in the oncological clinic are presented in Table 1. As a result of the research, it was revealed that out of 123 strains of microorganisms resistant to carbapenems, 88(71.5%) strains of bacteria were producers of carbapenemases. The carbapenemase-producing of type OXA-48 were: K.pneumoniae (86.5%), E.coli (9.1%), Enterobacter asburiae (100%) and Serratia marcescens (50.0%). The producers of NDM-type carbapenemases were also identified - this is Enterobacter cloacae (40.0%) and K.pneumoniae (2.7%). A. baumannii produced carbapenemases OXA-23 (97.1%) and OXA-40 (2.8%). Among the P.aeruginosa strains studied 46.1% produced VIM-type carbapenemases and 11.5% GES-5-like.
Conclusion
The species composition of gram-negative carbapenemresistant microorganisms isolated from the biological materials of cancer patients is presented as the most frequently isolated hospitals pathogens (A. baumannii, P. aeruginosa, E. coli, K.pneumoniae) and microorganisms, the relative amount of which as infectious complications pathogens not large (Enterobacter cloacae, Enterobacter asburiae, Serratia marcescens, etc.). Endemic strains of bacteria producers of globally common carbapenemases: OXA- 48, OXA-23, NDM, VIM - types are circulate in the oncological clinic. Obviously, regular monitoring of CarR strains of microorganisms, determination of resistance genes by molecular-genetic methods, well thought-out antimicrobial therapy and effective infection control strategy are necessary to effectively inhibit the spread of carbapenem-resistant bacteria.
References
- Dmitrieva NV, Petukhova IN, Grigorievskaya ZV, Dyakova SA, Sokolova EN, et al. (2018) Selected issues of epidemiology of resistant strains of gram-negative microorganisms Sepsis. Selected issues of diagnosis and treatment, ABC-press, pp. 416.
- Aginova VV, NV Dmitrieva, ZV Grigorievskaya, IN Petukhova, Bagirova NS, et al. (2017) Rational approaches to the treatment of nosocomial infections caused by gram-positive microorganisms in cancer patientsSiberian Oncological Journal 16(5): 12-17.
- CDC (2007) Healthcare Infection Control Practices Advisory Committee. Atlanta, GA: US Department of Health and Human Services, CDC, Healthcare Infection Control Practices Advisory Committee.
- Ageevec VA (2016) Molecular characterization of the producers of carbapenemases of the family Enterobacteriaceae, selected in St. Petersburg / dissertation for the degree of candidate of biological sciences. Ageevts VA, St. Petersburg. 137: 12.
- Nordmann P, Naas T, Poirel L (2011) Global spread of Carbapenemaseproducing Enterobacteriaceae. Emerging infectious diseases. 17(10): 1791-1798.
- Eidelstein MV, Sukhorukova MV, Skleenova EYu, Ivanchik NV, Mikotina AV, et al. (2017) Antibiotic resistance of nosocomial strains of Pseudomonas aeruginosa in hospitals in Russia: results of a multicenter epidemiological study “MARAFON” 2013-2014. Clinical microbiology and antimicrobial chemotherapy p. 37-41.
- Sukhorukova MV, Eidelstein MV, Skleenova E Yu, Ivanchik NV, Shek EA, et al. (2017) Antibiotic resistance of nosocomial Acinetobacter baumannii strains in hospitals in Russia: results of the multicultural epidemiological study “MARAFON” 2013-2014. Clinical Microbiology and Antimicrobial Chemotherapy, p. 42-48.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...




