Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-5945
Research Article(ISSN: 2638-5945) 
Application of Next Generation Sequencing Approach to Molecular Diagnosis Of Hereditary Colorectal Cancer: Identification Of A Novel Heterozygous Single Nucleotide Germline Deletion In Msh2 Gene Cause Lynch Syndrome Volume 3 - Issue 5
Zhe Lu1†, Lewei Yang2†, Santasree Banerjee3, Meifang Xiao1* and Yan Zhang4*
- 1Hainan Women and Children’s Medical Center, China
- 2Department of Radiation Oncology, The Fifth Affiliated Hospital of Sun Yat-sen University, China
- 3Department of Genetics, College of Basic Medical Sciences, Jilin University, China
- 4Department of Pathology, Shenzhen Longhua District Maternity & Child Healthcare Hospital, China
Received: May 14, 2020 Published: May 26, 2020
Corresponding author: Yan Zhang, Department of Pathology, Shenzhen Longhua District Maternity & Child Healthcare Hospital,
Shenzhen, China
Meifang Xiao, Clinical Laboratory, Maternal and Child Health Hospital of Hainan Province, Haikou, China
DOI: 10.32474/OAJOM.2020.03.000171
Abstract
Germline mutations in DNA mismatch repair (MMR) genes including MSH2, MLH1, MSH6 and PMS2 causes Lynch syndrome (LS). Lynch syndrome (LS) is the autosomal dominantly inherited familial form of colorectal cancer (CRC) predisposing syndrome. In our present study, we identified and screen a five generation Chinese family with LS. According to the Amsterdam II criteria, clinical diagnosis has been done for the affected members of this family. In this study, we performed genetic molecular study for 79 members (11 patients and 68 unaffected members) of this five generation Chinese family and 100 normal healthy control individuals of same ethnic background. Targeted next generation sequencing and Sanger sequencing identified a novel heterozygous single nucleotide germline deletion (c.1427delC) in MSH2 gene in the proband, which is co-segregated well with the disease phenotype among all the affected family members. This novel heterozygous deletion (c.1427delC) in MSH2 gene leads to the formation of a premature stop codon (p.Pro476Leufs*6) which finally results into the formation of a truncated MSH2 protein. Our present finding expands the mutation spectrum for the MSH2 gene as well as establishes the significance of targeted next generation sequencing for identifying novel candidate mutations for patients with Lynch syndrome.
Keywords: Lynch Syndrome; MSH2 Gene; Targeted Next Generation Sequencing; Novel Mutation; DNA Mismatch Repair Gene.
Introduction
LS is a colorectal cancer (CRC) predisposing syndrome with
an autosomal dominant mode of inheritance Lynch et al. [1]
approximately, 1-13% among all the colorectal cancer patients
has been suffering from LS primarily Vasen et al. [2]. The Clinical
symptoms or diagnostic features of LS are early onset CRC, along
with extracolonic manifestations; i.e. endometrial, pancreatic or
gastrointestinal cancers Canard et al. [3]. Clinical diagnosis for
the patients with LS has been done according to the Amsterdam
II criteria Canard et al. [3]. Identification of germline mutations of
DNA mismatch repair (MMR) genes and positive family history are
also significant factor for clinical diagnosis Chung et al. [4]; Goecke
et al. [5].
However, according to the previous reports, MSH2 was the
first gene to be identified as associated with LS, together with
MLH1 Knudsen et al. [6]. Germline mutations of MSH2 and MLH1
are together accounting for 90% cases of LS Peltomäki et al. [7].
During DNA replication, to repair the mismatched bases, the
MSH2 bind with MSH6 or MSH3 to form the MutSα/β complexes
and translocate into nucleus to bind to the DNA and start the
mismatched base repair Boland et al. [8]. Germline mutations in the DNA MMR genes cause formation of non-functional MMR protein
which in turn increases the rate of spontaneous somatic mutation.
Non-functional MMR protein associated somatic mutations are
majorly occurring in microsatellite sequences Woods et al. [9]. In
addition, till now, there were 500 germline mutations of DNA MMR
genes have been reported and 39% of these are MSH2 mutations
Vasen et al. [10].
Takeda et al [11] reported that germline loss-of-function
mutation in MSH2 gene also causes synchronous endometrial
and ovarian cancer (SEOC) in a 41-year-old Japanese patient with
a positive family history of colorectal and gastric cancers Takeda
et al. [11]. This patient was diagnosed with Lynch syndrome
and it is a rare example of a patient with genetically diagnosed
Lynch syndrome together with SEOC. SEOC is extremely rare and
caused by Lynch syndrome. In addition, LS patients with MSH2
and MLH1 germline mutations are identified with high-degree of
microsatellite instability Sanchez et al. [12]. Moreover, according
to the data deposited in International Society for Gastrointestinal
Hereditary Tumours (InSiGHT) database, among 3000 unique
germline mutations of MMR genes, MLH1, MSH2, MSH6 and PMS2
account for 40, 34, 18, and 8 %, respectively Thompson et al. [13].
Here, in order to understand the genetic and molecular basis
of the LS in this five generation Chinese pedigree, targeted nextgeneration
sequencing has been done with a panel of 14 genes
(APC, AXIN2, BMPR1A, EPCAM, MLH1, MLH3, MSH2, MSH6,
MUTYH, PMS1, PMS2, PTEN, SMAD4, STK11) associated with Lynch
syndrome. Targeted Next generation sequencing identified a novel
heterozygous single nucleotide germline deletion (c.1427delC,
p.Pro476Leufs*6) in MSH2 gene. Identified candidate variants by
next generation sequencing has been validated and confirmed by
Sanger sequencing. This germline deletion is co-segregated with LS
phenotype among all the affected family members, with autosomal
dominant mode of inheritance, in this five generation Chinese
family.
Results
Clinical Description
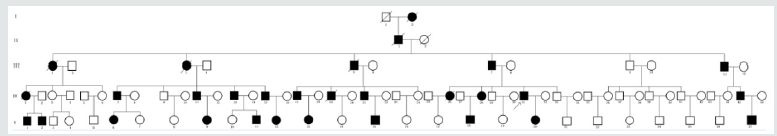
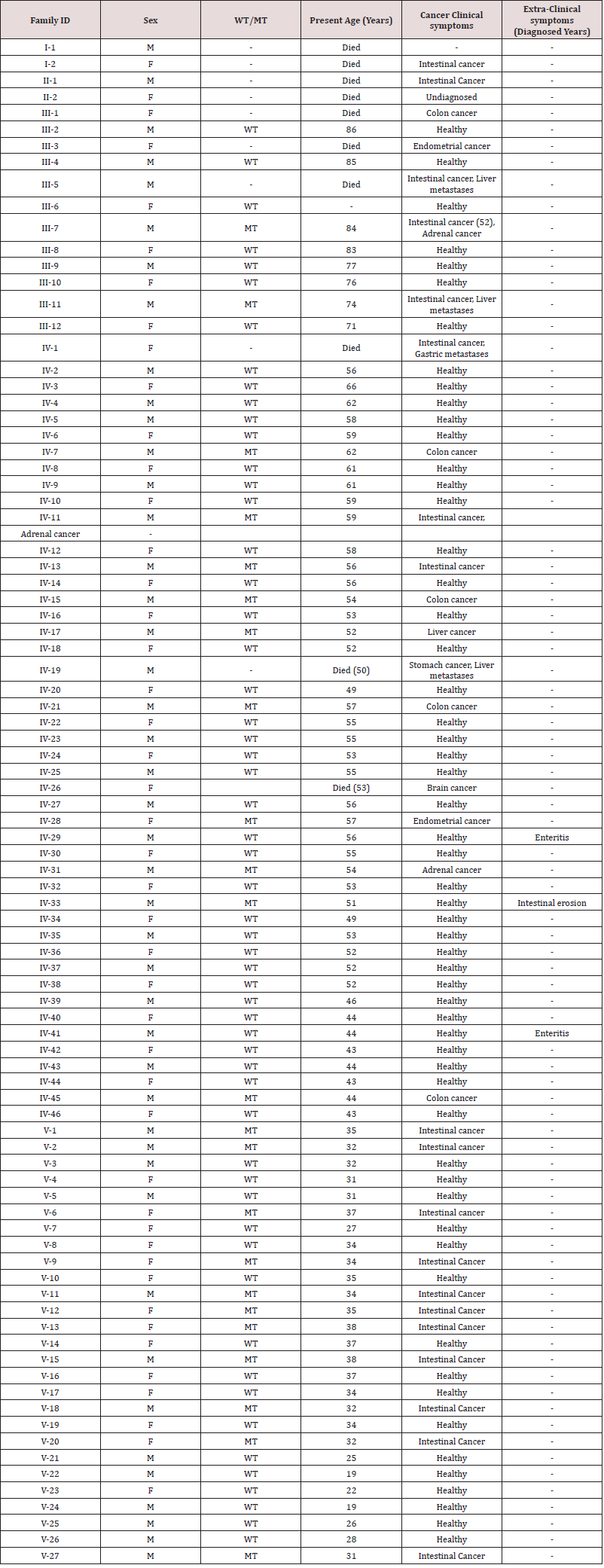
Family recruitment and clinical examination. We identified a five generation Chinese pedigree of 89 members (Figure 1) with LS. In (Table 1), we described the detailed clinical information for all the affected and unaffected members in this family.
Table 1: Clinical characteristics of all the affected and unaffected family members found in our study. [WT: Wild type; MT: Mutant type].
Identification and characterization of candidate mutation
Targeted next generation sequencing identified a heterozygous novel single nucleotide deletion; c.1427delC, p.Pro476Leufs*6 in MSH2 gene [NCBI Reference sequence NM_000251] in the proband (IV-31). Segregation analysis showed that this novel heterozygous deletion is co-segregated well with the LS phenotypes among the affected members, but not present in the unaffected members of this family. In addition, this novel deletion is also not present in the 100-normal control individual of same ethnic background.
Sanger Sequencing And Confirmation
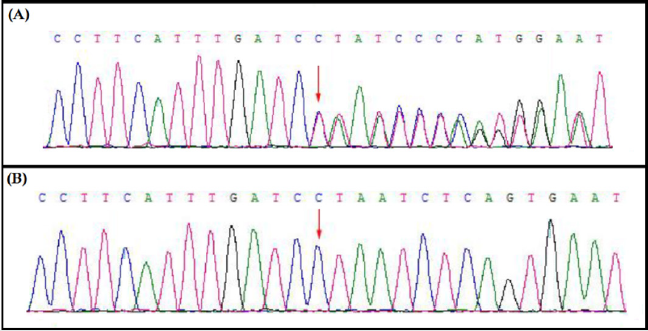
This novel heterozygous deletion; c.1427delC, p.Pro476Leufs*6 in MSH2 gene was confirmed by Sanger sequencing in proband and among all the affected family members in this family (Figure 2).
Discussion
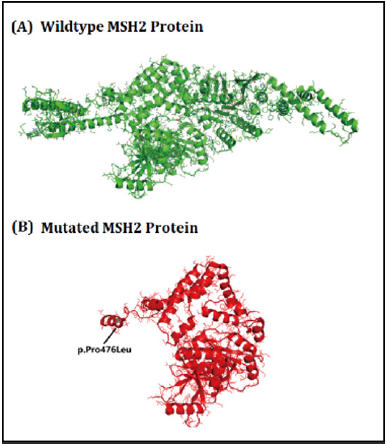
Here, we identified a five generation Chinese family with LS. After proper clinical diagnosis, genetics screening has been done by targeted next generation sequencing and confirmatory Sanger sequencing. Targeted next generation sequencing and Sanger sequencing identified a heterozygous novel single nucleotide deletion (c.1427delC, p.Pro476Leufs*6) [NCBI Reference sequence NM_000251] in the human MSH2 gene in the proband (IV-31) and in all the affected family members in this Chinese family. This heterozygous novel deletion of MSH2 gene has not presented in the ExAC database. This deletion finally results in the formation of truncated MSH2 protein (p.Pro476Leufs*6) by the formation of a premature termination codon (Figure 2). The structural effect of this mutation has been schematically showed in (Figure 3).
The function of DNA mismatch repair genes and its correlations with the human disease is very significant. Germline mutations in the human homologue of the bacteria mutS gene (MSH2) predispose individuals to Lynch syndrome Wang et al. [14]; Pérez-Cabornero et al. [15]. DNA mismatch repair (MMR) genes are following a highly conserved biological pathway playing a significant role in maintaining genomic stability. However, primary function of MMR gene is to repairing base-base mismatches and insertion/deletion mispairs generated during DNA replication and recombination. Furthermore, recently it has been reported that MMR gene also plays a key role in homologous recombination and in DNA damage signaling in eukaryotic cells. Germline truncated mutation of MMR leads to the formation of a non-functional MMR protein which finally results into genome-wide instability, increase the spontaneous mutation rate, predisposition to certain types of cancer including lynch syndrome, resistance to certain chemotherapeutic agents, and abnormalities in meiosis and sterility in mammalian systems Baglietto et al. [16].
According to recent reports it has been showed that LS patients with germline MSH2 mutations used to develop extracolonic cancers while LS patients with MLH1 mutations never develop extracolonic manifestations Lucía Pérez et al. [15]. In addition, LS patients with MSH6 mutations have the highest risk of developing endometrial cancer Lucía Pérez et al. [15]. In South-East Asia, LS patients are usually developed endometrial and stomach cancer Li [17]. In LS patients with MSH2 germline mutations, the lifetime risk of developing colorectal cancer is ranging between 57 % - 80 % Bonadona et al. [18]; Vasen et al. [10]. Lynch syndrome is caused by the germline mutations of MSH2 gene. Most of the reported mutations of MSH2 gene were leading to the formation of truncated MSH2 protein Li [17] ; Bakry et al. [19]. Presently, in world population, 5327 sequence variants of the MSH2 gene have been reported. Among all reported pathogenic mutations of the MSH2 gene, missense, nonsense and deletion mutations are the major types. In Chinese population, till now, 119 pathogenic mutations in MSH2 gene has been reported In conclusion, our present study identified a five generation Chinese family with LS. Proband and all the affected family members are harboring a heterozygous novel single nucleotide deletion in MSH2 gene. Our study not only expands the spectrum of the germline mutations of MSH2 gene in the Chinese population but also establish again the significance of targeted next generation sequencing for identifying candidate mutations in patients with LS. Moreover, our present finding also contributes to a database of germline mutations in MSH2 gene which is very significant in future for proper clinical diagnosis, easy molecular genetic screening patients with LS.
Materials and Methods
Ethical statement
Family members of this five generation Chinese family have given written informed consent as they are participating in this study. The Ethical Committee of the Maternal and Child Health Hospital of Hainan Province, Haikou, China, reviewed and approved our study protocol in compliance with the Helsinki declaration. Diagnosis of the patients for Lynch syndrome was made by oncologists, on the basis of Amsterdam II criteria.
Patients and pedigree
A five generation Chinese family with Lynch syndrome (Figure 1), diagnosed and treated in Maternal and Child Health Hospital of Hainan Province, Haikou, China, were enrolled in our study. The diagnostic standard or criteria for patients with Lynch syndrome was as follows: Diagnosis of Lynch syndrome is based on the Amsterdam criteria Canard et al. [20]. In this study, we performed genetic molecular study for 79 members (11 patients and 68 unaffected members) of this five generation Chinese family and 100 normal healthy control individuals of same ethnic background.
Targeted exome-based next-generation sequencing and variant identification
DNA samples obtained from the proband (IV-31) were sequenced using targeted next-generation sequencing. Roche NimbleGen’s (Madison,USA) custom Sequence Capture Human Array was used to designed to capture 98480 kb of targeted sequence, covering 181 exons and flanking sequence (including the 100 bp of introns) of 14 genes (APC, MLH1, MSH2, MSH6, PMS2, AXIN2, BMPR1A, EPCAM, MLH3, MUTYH, PMS1, PTEN, SMAD4, STK11) which is associated with colorectal cancer (CRC) and yielded an average of 6366534 reads per sample, with approximately 68.78% mapping to the targeted regions. The average sequencing depth of the target area is 464.68 with 99.46% coverage. The procedure for preparation of libraries was consistent with standard operating protocols. In each pooling batch, 10 to 33 samples were sequenced simultaneously on Illumina HiSeq 4000 Analyzers (Illumina, San Diego, USA) for 90 cycles (specially designed by us for this study). Image analysis, error estimation, and base calling were performed using Illumina Pipeline software (version 1.3.4) to generate raw data. The raw reads were screened to generate – clean reads‖ followed by established filtering criteria. Clean reads with a length of 90 bp were aligned to the reference human genome from the NCBI database (Build 37) using the Burrows Wheeler Aligner (BWA) Multi-Vision software package with output files in - bam‖ format. The bamdata were used for reads coverage in the target region and sequencing depth computation, SNP and INDEL calling, and CNV detection. First, a novel three-step computational frame work for CNV was applied. Then, SNPs and INDELs were called using SOAPsnp software and Sam tools pileup software, respectively. A SNP or INDEL would be filtered out if it could not follow the criterion: supported by at least 10 reads and >20% of the total reads. The frequency filter was set at 0.05. If a SNP frequency was more than 0.05 in any of the four databases (dbSNP, Hapmap, 1000 Genomes Project, the 124 healthy reference samples sequenced in this study), it would be regarded as a polymorphism, but not a causative mutation. Last, SNVs were retrieved in The Human Gene Mutation Database and the Leiden Open Variation Database and then labeled as reported or novel.
Sanger Sequencing
To validate the true positive of the mutation among all the family members (11 patients including the proband and 68 unaffected members), Sanger sequencing was performed. Primers flanking the candidate loci were designed based on the reference genomic sequences of Human Genome from GenBank in NCBI and synthesized by Invitrogen, Shanghai, China. PCR amplification was carried out in ABI 9700 Thermal Cycler. PCR products were directly sequenced on ABI PRISM 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA). Sequence data comparisons and analysis were performed by DNASTAR SeqMan (DNASTAR, Madison, Wisconsin, USA).
The novel heterozygous deletion identified in MSH2 gene through targeted next generation sequencing were verified through Sanger sequencing using the primers: F-5’- AAGGAGTTGTTCGTTTTCCACTT -3’, R-5’- TTACCAAAAGCCAGGTGACATTC -3’. The reference sequence NM_000251 of MSH2 was used.
Acknowledgments
We thankful to the proband and all the family members for their participation in this study.
Funding
There is no specific funding.
Availability Of Data And Materials
The data used to support the findings of this study are included within the article.
Authors’contributions
MX, YZ and SB designed the study. ZL performed the molecular diagnosis by Sanger sequencing. LY carried out the clinical analysis. YZ and MX contributed to the molecular diagnosis analysis based on NGS. ZL and LY wrote the paper. This project has been done under the guidance of ZL, MX, YZ and SB. All authors read and approved the final manuscript.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of Maternal and Child Health Hospital of Hainan Province, Haikou, China, and written informed consent was obtained from all participants.
Consent for publication
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare that they have no competing interests
References
- Lynch HT, Smyrk T (1996) Hereditary nonpolyposis colorectal cancer (Lynch syndrome). An updated review. Cancer 78(6): 1149-1167.
- Vasen HF, Watson P, Mecklin JP, Lynch HT (1999) New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116(6): 1453-1156.
- Canard G, Lefevre JH, Colas C, Coulet F, Svrcek M, et al. (2012) Screening for Lynch Syndrome in Colorectal Cancer: Are We Doing Enough? Ann Surg Oncol 19(3): 809-816.
- Chung DC, Rustgi, AK (2003) The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann Intern Med 138(7): 560-570.
- Goecke T, Schulmann K, Engel C, Holinski-Feder E, Pagenstecher C, et al. (2006) Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: a report by the German HNPCC Consortium. J Clin Oncol 24(6): 4285-4292.
- Knudsen NØ, Nielsen FC, Vinther L, Bertelsen R, Holten-Andersen S, et al. (2007) Nuclear localization of human DNA mismatch repair protein exonuclease 1 (hEXO1). Nucleic Acids Res 35(8): 2609-2619.
- Peltomäki P, Vasen H (2004) Mutations associated with HNPCC predisposition - Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers 20(4-5): 269-276.
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, et al. (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58(22): 5248-5257.
- Woods MO, Williams P, Careen A, Edwards L, Bartlett S, et al. (2007) A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat 28(7): 669-673.
- Vasen HF, Stormorken, Menko FH, Nagengast FM, Kleibeuker JH, et al. (2001) MSH2 mutation carriers are at higher risk of cancer than MLH1 mutation carriers: a study of hereditary nonpolyposis colorectal cancer families. J Clin Oncol 19(20): 4074-4080.
- Takeda T, Banno K, Yanokura M, Anko M, Kobayashi A, et al. (2018) Synchronous endometrial and ovarian cancer in Lynch syndrome with a MSH2 germline mutation: A case report. Mol Clin Oncol 9(5): 479-484.
- Sanchez de Abajo A, de la Hoya M, van Puijenbroek M, Godino J, Díaz-Rubio E, Morreau H, and Caldes, T. (2006) Dual role of LOH at MMR loci in hereditary non-polyposis colorectal cancer? Oncogene 25(14): 2124-2130.
- Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, et al. (2014) Application of a five-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants lodged on the InSiGHT locus-specific database. Nat Genet 46(2): 107-115.
- Wang J, Luo MH, Zhang ZX, Zhang PD, Jiang XL, et al. (2007) Clinical and molecular analysis of hereditary non-polyposis colorectal cancer in Chinese colorectal cancer patients. World J Gastroenterol 13(10): 1612-1617.
- Pérez-Cabornero L, Infante M, Velasco E, Lastra E, Miner C, et al. (2013) Genotype–phenotype correlation in MMR mutation-positive families with Lynch syndrome. Int J Colorectal Dis 28: 1195-1201.
- Baglietto L, Lindor NM, Dowty JG, White DM, Wagner A, et al. (2010) Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst 102(3): 193-201.
- Li GM (2008) Mechanisms and functions of DNA mismatch repair. Cell Res 18(1): 85-98.
- Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, et al. (2011) Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 305(22): 2304-2310.
- Bakry D, Aronson M, Durno C, Rimawi H, Farah R, et al. (2014) Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency consortium. Eur J Cancer 50(5): 987-996.
- Vasen HF, Möslein G, Alonso A, Bernstein I, Bertario L, et al. (2007) Guidelines for the clinical management of Lynch syndrome (hereditary nonpolyposis cancer). J Med Genet 44(6): 353-362.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...