Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2638-5945
Research Article(ISSN: 2638-5945) 
Analysis Of Mirna and Gene Expression Profiling In HPVAssociated Head and Neck Cancer Suggests New Scenarios Volume 5 - Issue 2
Mohsen Navari1-3, Maryam Etebari1-2, Navid Pourzardosht4, Arash Arashkia5, Davide Gibellini6, Pier Paolo Piccaluga7-8*
- 1Department of Medical Biotechnology, School of Paramedical Sciences, Torbat Heydariyeh University of Medical Sciences, Torbat Heydariyeh, Iran
- 2Research Center of Advanced Technologies in Medicine, Torbat Heydariyeh University of Medical Sciences, Torbat Heydariyeh, Iran
- 3Bioinformatics Research Group, Mashhad University of Medical Sciences, Mashhad, Iran
- 4Biochemistry Department, Guilan University of Medical Sciences, Rasht, Iran
- 5Department of Molecular Virology, Pasteur Institute of Iran, Tehran, Iran
- 6Department of Diagnostic and Public Health, University of Verona-Microbiology Unit, Verona, Italy
- 7Department of Medical Science and Surgery (DIMEC), University of Bologna, Bologna, Italy & Department of Pathology, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya
- 8Biobank of Research & Institute of Hematology and Medical Oncology Seràgnoli, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Italy
Received: December 23, 2022 Published: March 09, 2023
Corresponding author: Pier Paolo Piccaluga, Biobank of Research & Institute of Hematology and Medical Oncology “L. and A. Seràgnoli”, IRCCS Azienda Ospedaliero-Universitaria di Bologna; Department of Medical Science and Surgery (DIMEC), Bologna University Medical School, Bologna, Italy, Via Massarenti, 9 - 40138 Bologna, Italy.
DOI: 10.32474/OAJOM.2021.05.000212
Abstract
Human papillomavirus (HPV) is a risk factor for several human tumors, including head and neck cancers (HNC). The presence of the virus could impact the expression of the host microRNAs, leading to global change in the gene expression profile of the host cells. We studied such effects using in silico methods. We analyzed the gene expression profile of 38 HPV-positive and 180 HPVnegative HNC cases that were publicly available. Unsupervised and supervised hierarchical clustering and principal component analysis were performed to compare the two tumor categories. The deregulated genes were studied in terms of GO biological processes and KEGG pathways. The expression of the targets of the reported deregulated miRNAs was studied by GSEA. We found that HPV-positive and HPV-negative HNCs were similar, yet distinct entities, with the deregulated genes engaged mainly in cell cycle regulation and DNA repair. The targets of upregulated miRNAs in HPV-positive HNCs, surprisingly, were enriched in the same tumor category. We propose that this observation might be due to the different context of the tumor as compared to the cell lines used in the lab for miRNA target discovery, HPV interference with the cellular miRNA machinery, or due to the extremely high expression level of the miRNA targets. We report for the first time such a phenomenon, making further studies warranted.
Keywords: HPV: Human Papillomavirus; Gene Expression Profiling, MicroRNA, MicroRNA Expression Profiling
History
Human papillomavirus (HPV) is a DNA virus from the papillomavirus family comprising at least 170 types [1]. Several types of this virus, such as types 16 and 18, are engaged in the pathogenesis of some human cancers such as cervical and head and neck cancers (HNC) [2,3]. It is believed that the virus exerts such role through using its several oncoproteins, among which E6 and E7 are the leading players. Two important cellular tumor suppressors, i.e., pRB and p53, are the targets of these oncoproteins, with E6 interacting with p53 protein leading to its degradation in proteasome, and E7 competing with pRB binding, leading to cell cycle progression [4]. MicroRNAs (miRNAs) are a class of small RNAs with a post-transcription regulatory role on gene expression [5]. miRNAs have a fundamental role in the regulation of diverse biological processes, including carcinogenesis, to the extent that some have been designated as onco- and tumor-suppressor miRNAs [6]. The presence of viruses, including HPV, might impact the physiological balance of miRNA expression in the cell, and thus contribute to the pathological effects exerted by the virus, which would, in turn, lead to universal changes in the gene expression profile of the target cells [7]. Although some studies have dealt with gene and miRNA expression profiling in HNC based on the HPV association [8-12], however, to our best knowledge, an intercross between gene and miRNA expression profiling has not been reported so far. Thus, taking such an approach, we aimed to shed further light upon the role of deregulated human miRNAs in HPVassociated HNC.
Results
Gene expression profiling indicated that HPV-positive and HPV-negative HNC were similar yet distinct
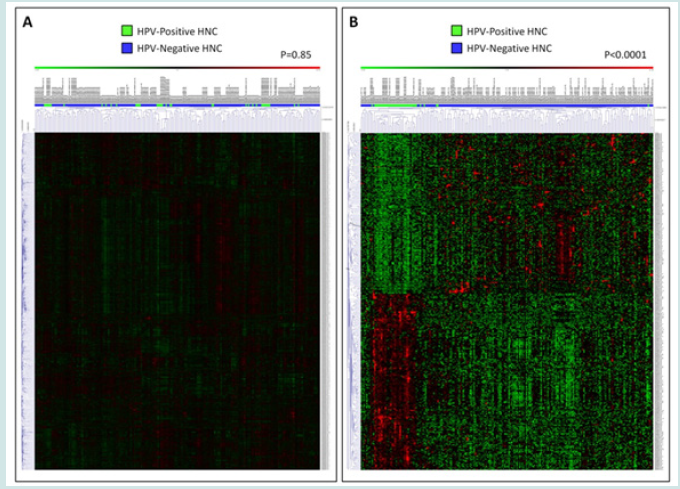
We first compared HNC cases based on HPV status. When unsupervised HCA and PCA methods were used, we found that the two groups were very similar. In fact, the two settings were intermixed in HCA (Fisher’s exact test p-value = 0.85, Figure 1A) and PCA (Cumulative Variance= 18.70%, Figure S1A). However, a t-test (FDR p < 0.05) and fold change (FC > 2) analysis indicated 201 and 181 probesets (173 and 145 unique genes) to be upregulated and downregulated in HPV-positive HNC, respectively (Table S1). The HCA analysis based on the deregulated genes could distinguish the two sets clearly (Fisher’s exact test p-value < 0.0001, Figure 1B), and PCA showed clustering of similar samples (Figure S2B, Cumulative Variance= 23.35%). This confirmed the existence of difference among the tumor cases based on the presence of the virus.
Figure 1: Hierarchical clustering analysis of gene expression profiling in head and neck cancer based on HPV status. (A) represents an unsupervised analysis, while (B) depicts the analysis based on genes differentially expressed between the two tumor categories.
Deregulated genes in HPV-positive HNC were mainly engaged in malignancy-related mechanisms
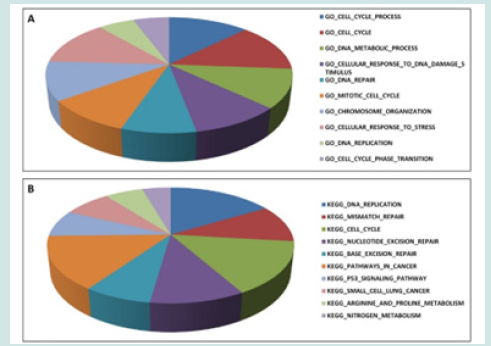
We then investigated to see what would be the function of the discovered deregulated genes in HPV-positive HNC. For this purpose, we used the MSigDB tool, looking at the Gene Ontology Biological Processes and KEGG pathways. Interestingly, processes such as cell cycle process, DNA metabolic process, DNA repair, response to DNA damage, chromosome organization, and DNA replication were found to be significantly enriched (Figure 2A). The enriched canonical pathways, again, mainly dealt with similar categories such as DNA replication, DNA repair, and p-53 pathway (Figure 2B).
Figure 2: Genes deregulated in HPV-positive HNC turned out to be implicated in malignancy-related processes. We used the MSigDB tool to analyze those genes based on Gene Ontology Biological Processes (A) and KEGG pathways (B).
Targets of both upregulated and downregulated miRNAs were enriched in HPV-positive HNC
Figure 3: GSEA reveals enrichment of the targets of the deregulated miRNAs in HPV-positive HNC in this tumor. The targets of both upregulated (A) and downregulated (B) miRNAs were enriched in HPV-positive HNC. Analysis of the targets of the individual miRNAs again showed enrichment of both categories in HPV-positive HNC (D-F).
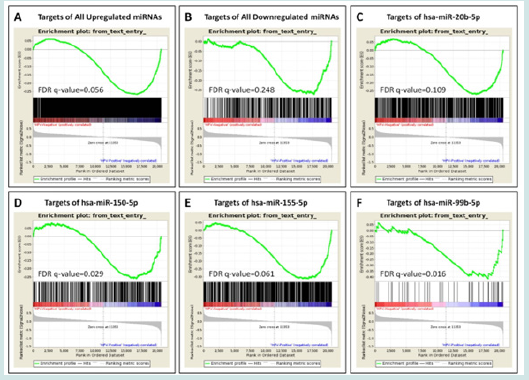
The miRNAs common between the two published researches included seven upregulated miRNAs (Hsa-miR-363-3p, Hsa-miR- 150-5p, hsa-miR-15a-5p, hsa-miR-20b-5p, Hsa-miR-625-5p, Hsa-miR-625-5p and hsa-miR-15b-5p) and two downregulated miRNAs (hsa-miR-99b-5p and Hsa-miR-744-5p) in HPV-positive HNC, respectively (Table 1). Based on MiRTarBase database, these microRNAs happened to target 3091 and 489 unique genes, for the induced and suppressed miRNAs, respectively (Table S2). When we analyzed the total targets of induced or suppressed miRNAs using GSEA, surprisingly, we found both of them enriched in HPV-positive HNC (FDR q-value 0.056 and 0.248, respectively, (Figure 3A-B). We then analyzed single miRNAs, and found 4 of them to be enriched in HPV-positive category: hsa-miR-20b-5p (FDR q-value= 0.109), hsamiR- 150-5p (FDR q-value= 0.029), hsa-miR-155-5p (FDR q-value= 0.061), and hsa-miR-
99b-5p(FDR q-value= 0.016) (Figure 3C-F).
Discussion
Several viruses are believed to be critical players in the development of some human malignancies [13]. Among them, HPV is one of the most studied viruses. This virus comprises numerous types, some of which like types 16 and 18 are engaged in the development of several cancers, including head and neck cancers [2]. The mechanisms by which HPV play such a role are diverse, but rely mainly on two virus-encoded E6 and E7 oncoproteins. The E6 protein is believed to interact with p53 protein, an essential tumorsuppressor protein, leading to its degradation in the proteasome. The E7 protein, on the other hand, competes with pRB binding, another tumor suppressor, leading to cell cycle progression [4].
The activity of viral oncoproteins could naturally lead to universal changes in the cells’ gene expression profile. One group of such targets that their expression could be affected, are miRNAs, small RNA molecules with a post-transcription regulatory role in gene expression [5]. Since their discovery, the role of miRNAs in different biological processes has been elucidated, including carcinogenesis [14]. Several miRNAs are considered as onco- and tumor-suppressor miRNAs [6]. It can be speculated that some part of the biological effects of HPV oncoproteins could be mediated through deregulated cellular miRNAs, which could affect the global gene expression profile of the tumor [15,16]. In this regard, and considering that HPV does not encode its miRNAs like some other viruses such as EBV [17], we looked at the role of such miRNAs on the global gene expression profile of HPV-associated head and neck cancers. We first looked at the gene expression profile of the HPVpositive and HPV-negative HNCs. The HCA and PCA analyses showed them to be very similar, being from the same origin. However, the supervised analysis showed them to be different in terms of the expression of some genes. Some studies have addressed this matter so far, and our results correspond to theirs [8-10]. However, we used a collection of all the cases presented in those studies, thus improving the accuracy of the findings.
The existence of deregulated genes in the two tumor settings would imply different mechanisms for the induction of malignancy. Thus, we analyzed the deregulated genes, and they turned out to be mainly engaged in two processes of cell cycle, and DNA metabolism and repair. This finding corresponds well to the role of E6 and E7, interfering with pRB and p53, the first being a significant cell cycle regulator, and the second an essential protein in DNA repair [4]. The analysis of the deregulated miRNAs targets, however, resulted in a surprising finding. Most of the targets of the upregulated miRNAs in the HPV-positive cancers were enriched in these cases, contrary to what expected. We re-ran the analysis to exclude the possibility of any errors. This phenomenon could be explained by several possibilities. First, the different cellular context of the experiments performed to determine the miRNA targets might have affected the results of the present study. Second, our observation might be an outcome of HPV interfering with miRNA machinery (as in the case of EBV [17]), thus impeding the physiological role of these miRNAs. Third, the expression levels of the targets of the induced miRNAs might be too high for the miRNAs to manipulate their expression significantly. In this point of view, the virus might use the miRNAs to fine-tune the over-expression of such genes. Of course, one could contest that the input data, i.e., the deregulated miRNAs used for the study might not be accurate. However, we used two different sources, choosing only the common miRNAs between the two, which were generated in tumor samples and, not in cell lines [11,12].
Altogether, we suggest for the first time that the role of deregulated miRNAs in the context of HPV, and more generally in the context of other viruses, might not be as expected. Thus, the researchers intending to use these miRNAs as potential targets of therapy shall consider more prudency.
Materials and Methods
We used four publicly available datasets which are available on GEO database (Gene Expression Omnibus of the National Center for Biotechnology Information-NCBI): GSE3292, GSE6791, GSE41613 and GSE53355, all generated on Affymetrix Human Genome U133 Plus 2.0 Array [8-10,18]. These included 218 cases, comprising 180 HPV-negative and 38 HPV-positive samples (Table S3). The data were normalized, as previously described [7,15,19]. In brief, the CEL files were imported into R version 3.2.3 and were normalized, and log2 transformed using RMA method in Bioconductor’s affy package version 1.48.0. In order to remove the batch effect among the four datasets, we used ComBat, setting the options to the default [20]. The data was further mean-variance normalized in geWorkbench version 2.6.0. Unsupervised and supervised hierarchical clustering analysis (HCA) were performed using MeV version 4.8.1 based on Pearson correlation and average linkage methods, the first being limited to the top 10,000 genes, due to the RAM restrictions. The p-value for the HCA accuracy was calculated by Fisher’s exact test using the online tool of GraphPad. Supervised and unsupervised principal component analysis (PCA) was also achieved using MeV software. The differentially expressed genes were determined using geWorkbench according to the following criteria: p ≤ 0.05 (adjusted Bonferroni false discovery rate (FDR)) and fold change ≥ 2. In order to assess the presence of relevant biological processes in the deregulated genes, we used Broad Institute’s MSigDB tool, looking at Gene Ontology Biological Processes and KEGG pathways (FDR q-value ≤ 0.50, top 10 enrichments chosen).
The miRNA signature of HPV in HNC was chosen from two previously published articles by Miller et al., 2015 and Lajer et al., 2012 [11,12]. Only the miRNAs common between the two datasets were chosen. To determine the targets of the deregulated miRNAs, we used MiRTarBase (Release 7.0: Sept. 15, 2017), a dataset which has collected the biochemically proven miRNA targets [21]). The expression of these targets was evaluated using gene set enrichment analysis software (GSEA) version 3.0, leaving the option to default [22].
Supplementary Materials
The followings are available online at www.mdpi.com/xxx/ s1, Figure S1: principal component analysis of gene expression profiling in head and neck cancer based on HPV status; TableS1: Genes deregulated in HNC based on the HPV status; Table S2: Targets of deregulated miRNAs extracted from MiRTarBase database; TableS3: Gene expression profiles used in this study.
Author Contributions
“conceptualization, PPP. and MN.; methodology, MN, NP, AA, and ME.; software, MN.; validation, MN, ME. formal analysis, MN.; resources, PPP.; writing-original draft preparation, MN and PPP.; writing—review and editing, MN, ME, PN, AA, and PPP.; supervision, AA and PPP.; funding acquisition, PPP”.
Conflicts of Interest
The authors declare no conflict of interest
References
- Bzhalava D, Guan P, Franceschi S, Dillner J, Clifford G (2013) A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 445(1-2): 224-231.
- Kobayashi K, Hisamatsu K, Suzui N, Hara A, Tomita H, et al. (2018) A Review of HPV-related head and neck cancer. Journal of clinical medicine 7(9): 241.
- Hildesheim A, Wang SS (2002) Host and viral genetics and risk of cervical cancer: a review. Virus research 89(2): 229-240.
- Hoppe-Seyler K, Bossler F, Braun JA, Herrmann AL Hoppe-Seyler F (2018) The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends in microbiology 26(2): 158-168.
- Testa U, Pelosi E, Castelli G, Labbaye C (2017) miR-146 and miR-155: two key modulators of immune response and tumor development. Non-Coding RNA 3(3): 22.
- Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO molecular medicine 4(3): 143-159.
- Navari M, Fuligni F, Laginestra MA, Etebari M, Ambrosio MR (2014) Molecular signature of Epstein Barr virus-positive Burkitt lymphoma and post-transplant lymphoproliferative disorder suggest different roles for Epstein Barr virus. Frontiers in microbiology 5: 728.
- Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, et al. (2006) Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res 12(3): 701-709.
- Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S et al. (2007) Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res 67(10): 4605-4619.
- Lohavanichbutr P, Mendez E, Holsinger FC, Rue TC, Zhang Y, et al. (2013) A 13-gene signature prognostic of HPV-negative OSCC: discovery and external validation. Clin Cancer Res 19(5): 1197-1203.
- Miller DL, Davis JW, Taylor KH, Johnson J, Shi, Z et al. (2015) Identification of a human papillomavirus–associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the cancer genome atlas. The American journal of pathology 185(3): 679-692.
- Lajer C, Garnaes E, Friis-Hansen L, Norrild B, Therkildsen, MH et al. (2012) The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. British journal of cancer 106(9): 1526.
- White MK, Pagano JS, Khalili K (2014) Viruses and human cancers: a long road of discovery of molecular paradigms. Clinical microbiology reviews 27(3): 463-481.
- Eastlack S, Alahari S (2015) MicroRNA and breast cancer: understanding pathogenesis, improving management. Non-Coding RNA 1(1): 17-43.
- Navari M, Etebari M, De Falco G, Ambrosio MR, Gibellini D et al. (2015) The presence of Epstein-Barr virus significantly impacts the transcriptional profile in immunodeficiency-associated Burkitt lymphoma. Frontiers in microbiology 6: 556.
- Mapekula L, Ramorola B, Hoosen TG, Mowla S (2019) The interplay between viruses & host microRNAs in cancer–An emerging role for HIV in oncogenesis. Critical reviews in oncology/hematology 137: 108-114.
- Navari M, Etebari M, Ibrahimi M, Leoncini L, Piccaluga P (2018) Pathobiologic roles of epstein–barr virus-encoded microRNAs in human lymphomas. International journal of molecular sciences 19(4): 1168.
- Parker HS, Leek JT, Favorov AV, Considine M, Xia X (2014) Preserving biological heterogeneity with a permuted surrogate variable analysis for genomics batch correction. Bioinformatics 30(19): 2757-2763.
- Piccaluga PP, Navari M, De Falco G, Ambrosio MR, Lazzi S, et al. (2016) Virus-encoded microRNA contributes to the molecular profile of EBV-positive Burkitt lymphomas. Oncotarget 7(1): 224.
- Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118-127.
- Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL et al. (2018) miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 46: D296-D302.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545-15550.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...