Lupine Publishers Group
Lupine Publishers
Menu
Research ArticleOpen Access 
Evaluation of Molecular Methods to Be Used in Sensors for The Detection of Bacteria in Water Volume 2 - Issue 3
Kotsiri Zoi, Panteleli Efstratia and Vantarakis Apostolos*
- *Department of Public Health, Medical School, University of Patras, Greece
Received: December 16, 2022 Published: December 23, 2022
Corresponding author: Apostolos Vantarakis, Department of Public Health, Medical School, University of Patras, Greece
DOI: 10.32474/JBRS.2022.02.000137
Abstract
Waterborne E. coli and E. faecalis remain a major economic problem worldwide because of their significant impact on health and disease. There is a constant need for new diagnostic tools that can detect low bacterial concentrations in a more cost- and timeeffective manner. The development of rapid and simple molecular detection in situ is required where specialized laboratory services are limited. Hereby, the recovery of DNA from artificially contaminated water samples of three different DNA extraction methods was investigated. Two commercial kits (DNeasy Ultraclean Microbial Kit, Dynabeads™ DNA DIRECT™ Universal Kit) and a boiling method were evaluated. All methods produced DNA in sufficient concentration ranging 64.55-184.7 ng/μL for E. coli and E. faecalis, respectively. Then, PCR methods were applied to confirm the effectiveness of the process. All amplifications had an LOD ranging from 102 to 101. Here is suggested that boiling extraction could be used as a method with rapid results, easy in use, adequate efficiency, which could be included in a portable sensor tool, as biosensor. The results of the present study highlight the importance of DNA extraction suitable for the application in a biosensor for a real time monitoring of the water samples.
Keywords: E coli; E faecalis; molecular detection; boiling method; rapid method; biosensors; PCR
Introduction
Pathogen contamination of environmental waters is a major health risk and a threat to future supplies of water for living organisms and their recreational activities. The consumption of unsafe water contaminated by bacteria, viruses, or parasites is responsible for waterborne diseases and is considered a major burden on public health, presenting a significant obstacle to socioeconomic development all over the world. For the determination of these health risks, seawater and drinking water using a series of tests for specific indicators which are defined in regional legislation need to be monitored [1]. The coastline represents a unique ecological system that is highly affected by industrial development and discharge of wastewater and reflects the water quality. Fecal contamination of seawater is considered an increase in concentration of Escherichia coli and intestinal enterococci. These aforementioned bacteria are present in the digestive system of mammals and birds but are also opportunistic pathogens for humans and animals [2]. E. coli and enterococci have been used as a standard for waterborne pathogens and are considered as fecal indicator bacteria in water quality testing all over the world because of their easy growth in cultural conditions [3].
The regular monitoring of E. coli and E. faecalis content in water permits avoiding health risks, especially from exposure in the probable context of short-term pollution on uncommon conditions. Moreover, drinking, bottled, drilling, springs, swimming pools, and marine waters were shown to be potential sources of resistant bacteria because they may contribute to the dissemination of microorganisms carrying antimicrobial resistance genes [4].
According to ISO 9308-1:2014/AMD 1:2016 (ISO 12016, n.d.), to estimate the quality of freshwater, the regulations require the total absence of E. coli and E. faecalis. For the monitoring of the microbiological quality of bathing and recreational seawater, the common routine practice is regular water sampling and analysis for E. coli and E. faecalis levels on a weekly or monthly basis.
European Directive 2006/7 / EC on bathing water quality management refers to “excellent quality” coastal water with less than 250 CFU for E. coli and 100 CFU for Enterococcus at 100 mL, “good quality” water up to 500 CFU for E. coli and 200 CFU for Enterococcus in 100 mL of seawater to be considered as “sufficient”. The assessment of the possible hazard depends on the detection methods that quantify bacterial indicator concentrations. Therefore, there is a requirement for an accurate and efficient method for the determination of water quality under stressed environmental conditions. Cell lysis and recovery of DNA is a prerequisite for PCR methodology. Cell lysis should be efficient and the presence of inhibitors in the eluted DNA should be limited.
Factors that may affect the efficiency of cell lysis may be the physiological characteristics of the species such as the structure of the cell wall, the physiological state of the cell (Coyne et al., 2004). As a result, most DNA extraction methods are ideal for one or a group of bacteria. So, a DNA extraction method that is optimal for all bacterial species should be chosen to overcome the limitations of a biosensor. Molecular detection methods based on isolation and amplification of nucleic acids such as PCR and isothermal amplification are capable of detecting the facial indicator bacteria in the short term and accurately [5]. Biosensors have raised enormous attention in the last decades. They are considered as powerful emerging tools for the detection of various biomarkers for both healthcare and environmental monitoring [6]. Biosensors can provide fast response in a short time, ultrasensitive detection of biomolecules, and have the potential to be miniaturized for portable use, requiring low volume of sample. Miniaturized based molecular methods face some limitations. The implementation of all experimental steps included such as centrifugation, spin columns etc. can be a hurdle. For instance, DNA extraction is considered to be the initial procedure as it requires specialized equipment. Recently, commercial kits have been developed to achieve quality and quantity of DNA although there is also boiling method that doesn’t require any complicated devices. The aim is to validate boiling in comparison with two commercially available kits that are widely used for DNA extraction. These two approaches have been applied as a control to evaluate the boiling with the standard methods. In this article, three extraction methods were evaluated in their efficiency in detecting E. coli and E. faecalis in water samples using three PCR methods to evaluate boiling as an extraction method in a biosensor.
Methods
Bacterial Strain and Growth Condition
Escherichia coli NCTC 9001 was reconstructed in Peptone Water (Merck, USA) for ten minutes at room temperature. Then, streak plate method was followed to obtain a single colony on a selective medium, Chromocult® Coliform Agar (CCA) (Merck, USA), at 37°C for 24 hours. E. faecalis NCTC 12697 was incubated on this Slanetz and Bartley (SB) agar (Oxoid, England) at 37°C for 48 hours. After the incubation, a single colony was collected, and the inoculum was transferred into 30 ml of Tryptone Soy Broth (TSB) (Oxoid, England). and incubated at 37° for 16 to 18 hours by shaking at 160 rpm. Following the procedure for bacterial determination, the optical density was measured using a photometer at 600 nm. Water samples were spiked with pure cultures of E. coli and E. faecalis in the range of approximately 109 CFU/mL. Serial dilutions (10-1-10- 8) were performed, and bacterial concentrations were counted in petri dishes. Glycerol at ratio 4:1 was added into samples to obtain stock cultures.
DNA extraction
Genomic DNA was extracted in three ways. Bacterial stocks were centrifuged at 4000 rpm for 5 min and then washed and suspended with 1 mL dH2O in duplicates, for each organism. The two 1.5 mL tubes containing E. coli and E. faecalis, genomic DNA were extracted, using the DNeasy Ultraclean Microbial Kit (Qiagen, Germany) according to the manufacturer’s instructions. In the other two tubes, DNA extraction was performed by applying the Dynabeads™ DNA DIRECT™ Universal Kit (Thermo Fisher, Waltham, MA, USA) following the manufacturer’s protocol. The last extraction method was achieved by adding 100 μL dH2O in bacteria pellets’ and then the samples were heated at 100oC for 10 minutes. To quantify the amount of DNA, the samples were verified spectrophotometrically using a NanoDrop 1000 instrument (Thermo Scientific, Wilmington, DE, USA). Then, serial dilutions were obtained, and PCRs were followed. Experiments were performed in triplicate.
Colorimetric LAMP
The primers for the LAMP reactions of the E. coli and E. faecalis strains were designed to target the region of the b-D-glucuronidase (uidA) [7] and Enterococcus 23s rRNA genes (Lee et al., 2019). A set of six primers, two outer primers, a forward outer primer (F3) and a backward outer primer (B3); two inner primers, a forward inner primer (FIP) and a backward inner primer (BIP), and two loop primers (LF and LB) (Eurofins, Germany). The LAMP assay was performed in a total 15 μl mixture containing WarmStart® Colorimetric LAMP 2X Master Mix (New England Biolabs, Singapore), 10x LAMP primer mix (1.6 mM FIP and BIP, 0.2 mM F3 and B3, 0.4 mM of the LF and LB), 1 μL genomic DNA and sterile deionized water, according to the manufacturer’s instructions. The LAMP reactions were carried out at 64oC for 60 min in triplicate.
PCR
To validate the heating-based DNA extraction method, water samples spiked with E. coli and E. faecalis were evaluated in parallel by commercial DNA extraction kits. Conventional PCR was performed with two outer LAMP primers, forward outer primer (F3) and backward outer primer (B3). All reactions were performed on a MJ Mini Personal Thermal Cycler (Bio-Rad, USA). The PCR reaction mixture had a total volume of 20 μL and consisted of 10 x Dream Taq PCR buffer (Thermo Fisher Scientific, USA), 0.2 mM dNTPs, 0,6 mM betaine, 1.25 U of Dream Taq DNA polymerase, 0,25 uM forward and reverse primers, template DNA, and nuclease-free water. All reactions were performed on tubes and flat caps strips of 8. (Thermo Fisher Scientific, USA), and the cycling conditions for PCR with target E. coli were carried out under the following program: initial denaturation at 95oC for 3 min; followed by 35 cycles of 95oC for 30 s, annealing at 55oC for 30 s, extension at 72oC for 30 and finally 72oC for 10 min. For E. faecalis, the cycling conditions for PCR were initial denaturation at 95oC for 5 min; followed by 35 cycles of 95oC for 30 s, annealing at 52oC for 30 s, extension at 72oC for 1 min, and finally 72oC for 5 min. The amplified products were analyzed by gel electrophoresis in 2 % ultrapure agarose gels (AgaPure™ Agarose LE, Canvax, Biotech) by addition of TAE buffer at 100 V for 1 hour. For the visualization of DNA bands, UV light with a camera on it was used to determine the results. The loaded PCR products on 2 % agarose gel obtained fragment’s size about 253 bp for E. coli and 407 bp E. faecalis. The procedure followed in triplicate.
Quantitative PCR (qPCR)
The evaluation was carried out by comparing three calibration curves using the DNA extracted by commercial kit methods and boiling, diluted with purified water. Quantitative PCR amplification was performed using the Agilent AriaMx Real-Time PCR System (Agilent Technologies, USA). To compare the DNA extraction methods, two outer LAMP primers (F3 and B3) were used. All qrtPCR reactions were performed in a total volume of 20 μl, consisting of 2 x KAPA SYBR® FAST qPCR Master Mix (2X) Kit (Sigma-Aldrich, USA), 0,2 uM forward and reverse primers, template DNA and nuclease-free water. The qPCR reaction conditions were as follows for E. coli: 95oC for 3 min, followed by 40 cycles of 95oC for 3 sec, 58oC for 20 sec and 72oC for 7 sec. The conditions were as follows for E. faecalis were 95oC for 3 min, followed by 40 cycles of 95oC for 10 sec, 56oC for 20 sec and 72oC for 5 sec. Each dilution was analyzed in duplicate.
Statistical Analysis
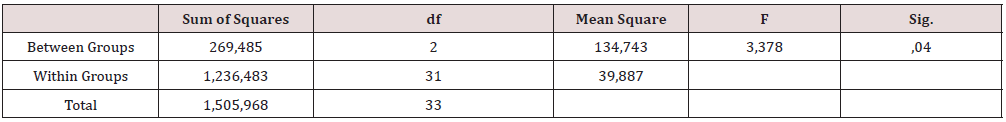
Statistical analyses were performed with the IBM SPSS Version 27.0 statistical software package and Microsoft Excel to compare the extraction methods of each microorganism. A significance level was adopted (p<0.05). The comparative analysis of the Ct values for both the target region of the uidA and Enterococcus 23s rRNA genes obtained in the amplification of serial dilutions of DNA extracted with three methods was performed using ANOVA (α = 0.05).
Results & Discussion
Numerous methodologies could be used for the extraction of DNA from microbial samples. These strategies are focusing on enzymatic, chemical, or thermal lysis, mechanical disruption of the cell wall by beads or sonication, or a combination of the above [8]. In this study, we applied three different extraction technologies requiring separated laboratory equipment to test their efficiency, sensitivity, and rapidness in extracting amplifiable E. coli and E. faecalis DNA from water samples. The quantity and quality of nucleic acids are described below. The Ultraclean microbial kit includes two ways for DNA collection, mechanical disruption where microorganisms are lysed by a combination of heat, detergent and mechanical force against specialized beads and column-based purification silica gel. Another advantage is the simple transport and storage conditions that can be kept at room temperature. The Dynabeads concentrate DNA by the conjugation of genomic material with magnetic beads and the boiling one is a simplified DNA extraction method with reduced time analysis and no requirements of specialized laboratory equipment. It consists of a lysis buffer with magnetic beads and, in addition, requires a magnetic stand. Dynabeads makes use of only one solution and washing steps and, as such, occupies more volume and weight than the other method which is proportional to sample. Similarly, the last method for DNA extraction purposes was performed by boiling in a water bath being cost effective and efficient.
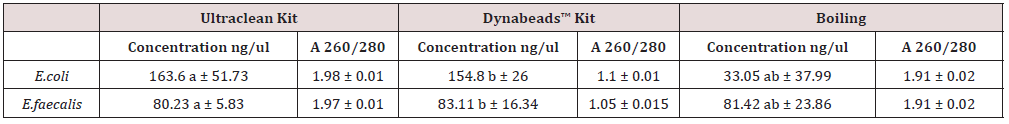
Among them, the boiling method is not only superior in terms of simplicity, cost effectiveness, and time of the process, but it is also consistent, based on its low bias [9]. Performance of the boiling method has been also demonstrated for fecal microbes [10] and human oral microbes [11], suggesting that it could be widely reliable for other microorganisms. In our study, both microbial kit and Dynabeads were, indeed, fast, and easy to use by single-step isolation procedures and processing times of 40 and 15 min for each process run of samples, respectively. DNA extraction methods with comparable processing times have been reported previously [12]. The protocol of two kits, according to manufacturer’s instructions, both need low initial volume. The sample volume was determined to be 1 mL. As it is concerned the elution volume ranged from 50 μL to 1000 μL. The ratio of absorbance at 260 nm and 280 nm is used to evaluate the purity of DNA. A 260/280 ratio by Nanodrop from initially extracted DNA samples ~1.8 is generally accepted as “pure” for DNA. If the ratio is appreciably lower in either case, it may indicate the presence of protein or other contaminants that absorb strongly at or near 280 nm. In the current study, it was found to be in a range of 1.8-2 for Ultraclean microbial kit, 1.8-2 for Dynabeads and 1.05-1.19 for boiling method. By referring to concentration values, those ones were measured 73.5-158 ng/μL, 7.15-98.3 ng/ μL and 64.55-184.7 ng/μL, respectively (Table 1).
It is well known that spin column DNA extraction method can generate high purity DNA. Extraction is a demanding process that includes several steps, and it needs to be optimized to minimize the danger of DNA loss in each manipulation [13]. Nevertheless, in this study, the extraction efficiency of the spin column extraction method is estimated to be the highest. The 1 ml initial water samples contain 108 bacteria. After the extraction was followed and then serial dilutions were performed in tubes containing separately the two types of bacteria. Molecular approaches have been progressively used for the detection and quantification of risky microorganisms [14]. The need for a fast and easily applicable in situ molecular detection and quantification method is essential for gaining information for environmental analysis. Another critical step for the detection is the nucleic-acid-based quantification of bacteria, as part of sample processing foregoing to the real measurement, as has previously been mentioned [15,16]. The results of this study verify that DNA extraction is a critical step of this process. Extraction enables the selective removal of components in a mixture in order to minimize the inhibiting factors. Interestingly, a significantly higher quantity of DNA was extracted using the Ultraclean kit, possibly due to its use of mechanical disruption and filtration column with a silica membrane, compared to the other kits that employed magnetic beads as it was observed in E. coli.
The magnetic bead-based system needs more time and surface area for binding nucleic acids than the silica column-based method. Nevertheless, there is disagreement on which of the two systems offers higher nucleic acid yield [17]. The results of the present study showed that all DNA extraction methods that have been examined yielded nucleic acid levels sufficient for DNA amplification. In terms of the quality of extracted DNA, the samples had relatively pure nucleic acids, as assessed by the 260/280 absorbance ratio, as it is concerned commercial kits. Studies have proposed as a more efficient method of DNA extraction the boiling method than other in-house methods that are time-consuming and require high quantities of inputs such as microwave heating and shaking with pure phenol for Gram-positive and Gram-negative bacteria. [18,19]. Most importantly, this boiling-based DNA extraction method achieved a high quality of DNA extraction with rapidity and simplicity without any specified apparatus such as a centrifuge or vortex. Use of PCR has provided an efficient means of amplifying samples with boiling method for testing environmental samples (Tables 2&3). Studies showed similar or higher results than commercial kit methods [20]. Direct boiling using certain amounts of feces could significantly reduce the cost of DNA extraction and improve the efficiency and reduce sample preparation for the DNA extraction of facial microorganisms.
Table 2: Cycles threshold accompanied with standard deviations values for the applied DNA extraction methods for E. coli.
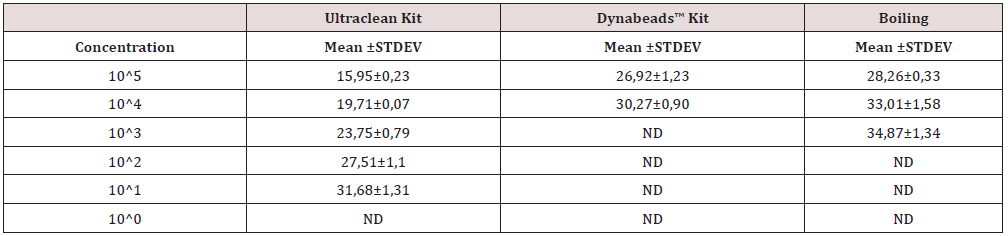
Table 3: Cycles threshold accompanied with standard deviations values for the applied DNA extraction methods for E. Faecalis.
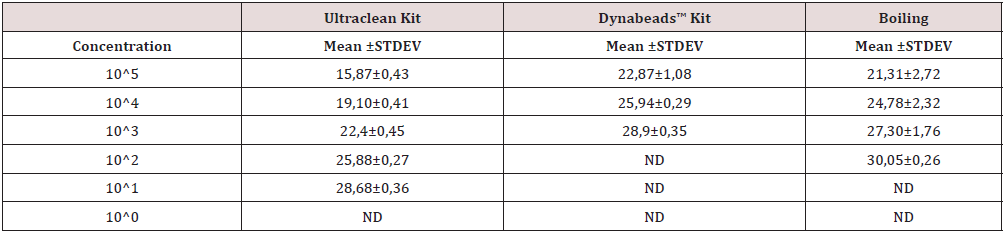
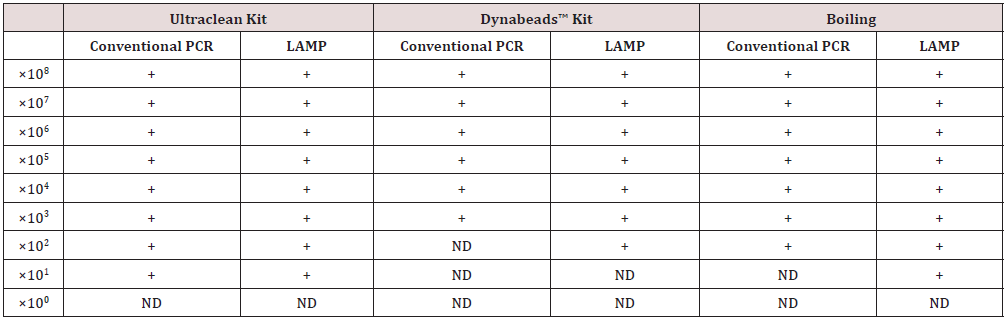
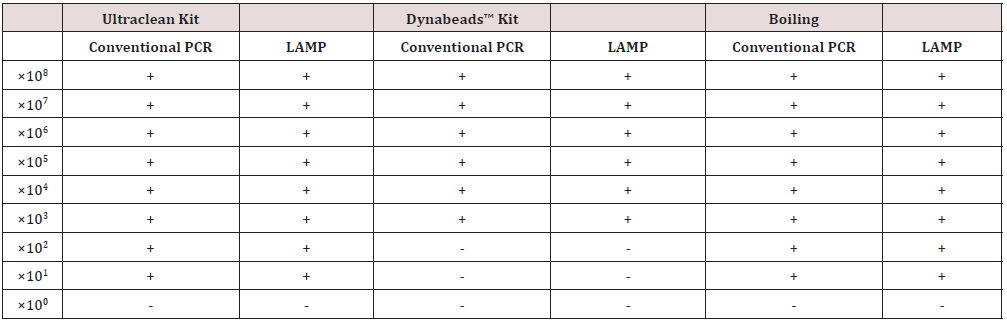
All DNA extraction methods resulted in similar results in downstream PCR applications. DNA dilutions were obtained by serial dilutions of an initial sample 108 cfu/ml. The LAMP PCR confirmed that boiling-based method successfully extracted DNA up to 101 CFU/mL for E. coli and E. faecalis. For the E. coli samples, 1 out 3 samples were positive in 101 CFU/mL, suggesting that DNA extraction efficiency of gram- negative bacteria was low due to cell walls with thick layers of peptidoglycan. The PCR confirmed that the heating-based method successfully extracted DNA up to 102 CFU/mL of E. coli and E. faecalis, respectively. As for the microbial kit, amplification product was detected even at template dilution corresponding to approximately up to 101 CFU/mL of E. coli and up to 100 CFU/mL for E. faecalis. With the Dynabeads kit, the lower detected DNA was up to 102 CFU/mL of E. coli and up to 100 CFU/ mL for E. faecalis. Also, the 10-fold serial dilutions of genomic DNA of the E. coli and E. faecalis were used to estimate the DNA extraction efficacy by a conventional PCR using two outer LAMP primers F3 and B3. LAMP colorimetric method, compared to qPCR, has been demonstrated as a sensitivity of 93.75% and specificity of 100% (Table 4) from samples of a breeding by boiling DNA extraction [21].
Table 4: Comparing the sensitivity of the Real time PCR assay on water extracts purified by the three DNA purification methods, spiked with E. coli and E. Faecalis.

As we see, there are many methods that suggest boiling methods for the detection of bacteria [22], helminthes [23], cysts and dinoflagellates [24]. Some of them combine the boiling method with the amplification with LAMP method for the detection of malaria diagnosis at the point of care [25]. Other study used boiling method and colorimetric LAMP for the rapid analysis and the limit of detection was counted about 2.4 - 3.7 parasites/μL [26]. Grampositive microorganisms can easily have their cell walls disrupted by boiling [27]. This may be related to the composition of the cell wall of Gram-positive microorganisms, which has peptidoglycan responsible for increased rigidity to the wall of Gram-negative microorganisms [28]. In the current study, boiling was sufficient for DNA extraction and PCR amplification in Gram-positive bacteria. As the method is fast, cheap, and easy to perform, the boiling of suspensions is an effective method for molecular studies of these microorganisms. However, several studies confirm that boiling method has already been applied in a huge variety of molecular methods, such as NGS, PCR, qPCR, etc [29]. Nevertheless, further studies are important in order to evaluate the method for more environmental matrices. The analyses were performed using the KAPA SYBR® FAST qPCR Master Mix on the Agilent AriaMx Real- Time. Analytical sensitivity of the RT-qPCR assay was assessed by determining the LOD for each gene using plasmid DNA containing the b-D-glucuronidase (uidA) and Enterococcus 23s rRNA genes and they showed variances in window amplification among the extraction methods.
The results presented in Tables 5&6 show that the analyses with Ultraclean Microbial kit had a mean LOD of 101 cfu in E.coli and in E. Faecalis. Dynabeads were less sensitive up to 104 cfu in E.coli and 103 cfu in E. Faecalis and boiling methods could detect organisms up to 103 cfu in E.coli and 102 cfu in E. Faecalis in triplicate runs. The elution volume was 50 ul in commercial kits and in boiling method about 1 ml. This can explain the higher LOD among the results in water samples. It is evident that those methods could also be used in a portable apparatus such biosensor because they don’t demand any special equipment. Similar devices have been suggested for applications in the field of food safety, such as in the detection of foodborne pathogens, allergens and genetically modified organisms to food by the usage of LAMP method [30]. Knowing this and the variety of its applications, it could be used in the field for the determination of fecal pollution in the environment. The comparative analysis of the Ct values for the b-D-glucuronidase and the Enterococcus 23s rRNA targets obtained in the amplification of serial dilutions of DNA extracted with three extraction methods was performed using ANOVA (α = 0.05) and showed statistically significant differences between the analytical curves of each target (p=0.04) (Table 7). The effect of three DNA extractions methods on the real-time PCR efficiency quantification was studied by [31]. The amplification efficiency calculated from the Cq values were all between 85 and 105% for E.coli and 104- 123% for E. Faecalis.
Conclusions
The results in the DNA extraction by boiling that is proposed in this study was as efficient as the commercial kits for the PCR and PCR LAMP detection of uidA and 23S rRNA genes of the microorganisms which were evaluated. By focusing on the processes by which boiling method results arise, this method for bacteria is considered as an important alternative for carrying out molecular studies since it is as efficient as the commercial kit but much less costly and laborious. It provides a platform of sample-to-answer results in less than 30 min. The suggestion is considered fit for purpose as an analytically validated mobile LAMP PCR permits experts to monitor specific genetic material in their samples as they investigate for pathogens affecting dynamic ecosystem or test fast the changes in water quality. This would permit a complete portable PCR system to be a user-friendly platform with easily sharing results.
Acknowledgments
This work was supported by the Single State Action Aid for Research, Technological Development & Innovation “INVESTIGATE - CREATE – INNOVATE” project “SMART-SEATRAC (No, T1EDK-04615)”.
References
- Ahmad F, Stedtfeld RD, Waseem H, Williams MR, Cupples AM, et al. (2017) Most probable number - loop mediated isothermal amplification (MPN-LAMP) for quantifying waterborne pathogens in < 25 min. J Microbiol Methods 132: 27-33.
- Bastien P, Procop GW, Reischl U (2008) Quantitative real-time PCR is not more sensitive than “conventional” PCR. J Clin Microbiol 46(6): 1897-1900.
- Baümler AJ, Sperandio V (2016) Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535(7610): 85-93.
- Biswal D (2016) Advances in Loop-Mediated Isothermal Amplification (LAMP) Technology and its Necessity to Detect Helminth Infections: An Overview. Biomarkers J 2: 1-6.
- Coyne SR, Craw PD, Norwood DA, Ulrich MP (2004) Comparative analysis of the Schleicher and Schnell IsoCode Stix DNA isolation device and the Qiagen QIAamp DNA Mini Kit. J Clin Microbiol 42(10): 4859-4862.
- Da Costa Andrade V, Del Busso Zampieri B, Ballesteros ER, Pinto AB, Fernandes Cardoso de Oliveira AJ (2015) Densities and antimicrobial resistance of Escherichia coli isolated from marine waters and beach sands. Environ Monit Assess 187(6): 1-10.
- Dilhari A, Sampath A, Gunasekara C, Fernando N, Weerasekara D, et al. (2017) Evaluation of the impact of six different DNA extraction methods for the representation of the microbial community associated with human chronic wound infections using a gel-based DNA profiling method. AMB Express 7(1): 179.
- Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, et al. (2006) Erratum: Real-time PCR in clinical microbiology: Applications for routine laboratory testing. Clin Microbiol Rev 19(1): 165-256.
- Fuquay JW, McSweeney PL, Fox PF (2011) Encyclopedia of dairy sciences.
- Huang T, Li L, Liu X, Chen Q, Fang X, et al. (2020) Loop-mediated isothermal amplification technique: Principle, development and wide application in food safety. Anal Methods 12(46): 5551-5561.
- Water quality — Enumeration of Escherichia coli and coliform bacteria — Part 1: Membrane filtration method for waters with low bacterial background flora — Amendment 1. ISO.
- Karle M, Miwa J, Czilwik G, Auwärter V, Roth G, et al. (2010) Continuous microfluidic DNA extraction using phase-transfer magnetophoresis. Lab Chip 10(23): 3284-3290.
- Kim JH, Jung S, Oh SW (2020) Combination of bacteria concentration and DNA concentration for rapid detection of E. coli O157:H7, L. monocytogenes, and S. Typhimurium without microbial enrichment. Lwt 117(4): 108609.
- Kotsiri Z, Vantarakis A, Rizzotto F, Kavanaugh D, Ramarao N, et al. (2019) Sensitive detection of E. coli in artificial seawater by aptamer-coated magnetic beads and direct PCR. Appl Sci 9(24): 5392.
- Lee S, Khoo VSL, Medriano CAD, Lee T, Park SY, et al. (2019) Rapid and in-situ detection of fecal indicator bacteria in water using simple DNA extraction and portable loop-mediated isothermal amplification (LAMP)PCR methods. Water Res 160: 371-379.
- Lim MY, Song EJ, Kim SH, Lee J, Nam Y Do (2018) Comparison of DNA extraction methods for human gut microbial community profiling. Syst Appl Microbiol 41(2): 151-157.
- Martins FH, Guth BEC, Piazza RM, Leão SC, Ludovico A, et al. (2015) Diversity of Shiga toxin-producing Escherichia coli in sheep flocks of Paraná State, southern Brazil. Vet Microbiol 175(1): 150-156.
- Mohd Nosi MZ, Syed Jamil Fadaak SNE, Muhammad MDD, Iehata S (2018) Assessment of gut microbiota in different developmental stages of Malaysian Mahseer (Tor tambroides). Aquac Res 49(1): 2977-2987.
- Nagai S, Yamamoto K, Hata N, Itakura S (2012) Study of DNA extraction methods for use in loop-mediated isothermal amplification detection of single resting cysts in the toxic dinoflagellates Alexandrium tamarense and A. catenella. Mar Genomics 7: 51-56.
- Nolasco O, Montoya J, Rosales Rosas AL, Barrientos S, Rosanas-Urgell A, et al. (2021) Multicopy targets for Plasmodium vivax and Plasmodium falciparum detection by colorimetric LAMP. Malar J 20(1): 225.
- One Health. WHO.
- Pandey PK, Kass PH, Soupir ML, Biswas S, Singh VP (2014) Contamination of water resources by pathogenic bacteria. AMB Express 4: 51.
- Peng X, Yu KQ, Deng GH, Jiang YX, Wang Y, et al. (2013) Comparison of direct boiling method with commercial kits for extracting fecal microbiome DNA by Illumina sequencing of 16S rRNA tags. J Microbiol Methods 95(3): 455-462.
- Perera RS, Ding XC, Tully F, Oliver J, Bright N, et al. (2017) Development and clinical performance of high throughput loop-mediated isothermal amplification for detection of malaria. PLoS One 12(2): e0171126.
- Rantakokko-Jalava K, Javala J (2010) Optimal DNA isolation method for detection of bacteria in clinical specimens by using the technique of PCR. Pakistan. J Clin Microbiol 40(11): 4211-4217.
- Ribeiro JC, Tamanini R, Soares BF, De Oliveira AM, De Godoi Silva F, et al. (2016) Efficiency of boiling and four other methods for genomic DNA extraction of deteriorating spore-forming bacteria from milk. Semin Agrar 37(5): 3069-3078.
- Sheikh N, Kumar S, Sharma HK, Bhagyawant SS, Thavaselvam D (2020) Development of a Rapid and Sensitive Colorimetric Loop-Mediated Isothermal Amplification Assay: A Novel Technology for the Detection of Coxiella burnetii From Minimally Processed Clinical Samples. Front Cell Infect Microbiol 10: 127.
- Sowmya N, Thakur MS, Manonmani HK (2012) Rapid and simple DNA extraction method for the detection of enterotoxigenic Staphylococcus aureus directly from food samples: Comparison of PCR and LAMP methods. J Appl Microbiol 113(1): 106-113.
- Tongeren SP Van, Degener JE, Harmsen HJM (2011) Comparison of three rapid and easy bacterial DNA extraction methods for use with quantitative real-time PCR. Eur J Clin Microbiol Infect Dis 30(9): 1053-1061.
- Xiong X, Huang M, Yuan F, Lu L, Xiong, X (2019) Development and Validation of a Fast DNA Extraction Protocol for Fish Products. Food Anal Methods 12: 1998-2008.
- Yamagishi J, Sato Y, Shinozaki N, Ye B, Tsuboi A, et al. (2016) Comparison of boiling and robotics automation method in DNA extraction for metagenomic sequencing of human oral microbes. PLoS One 11(4): e0154389.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...