Lupine Publishers Group
Lupine Publishers
Menu
Research ArticleOpen Access 
Automated Biosensor for Rapid Field Detection of Fecal Biomarkers in Coastal Areas Volume 2 - Issue 4
Kotsiri Zoi1, Panayotou Panayotis2, Beskos Panayotis2, Vantarakis Apostolos1*
- 1Department of Public Health, Medical School, University of Patras, 26504 Greece
- 2Metricon S.A., 26504, Rio, Greece
Received:February 12, 2024; Published: February 21, 2024
Corresponding author: Vantarakis Apostolos, Department of Public Health, Medical School, University of Patras, 26504 Greece
DOI: 10.32474/JBRS.2024.02.000145
Abstract
This study was conducted to introduce and assess a novel automated system for monitoring of Fecal Indicator Bacteria and therefore evaluate seawater quality. The system presents the comparative advantage that it can give accurate results in real time and in the field. Here, we develop the seawater analyzer and evaluate the process by comparing the function of the device with plate count technique known as gold standard method. We demonstrate a system that changes color with the bacterial presence and is considered to be accurate, rapid and precise to warn for fecal contamination. We also present results by applying the method in different seacoasts regarding the swimming activity during the winter months but also the activity that concerns only at the beginning of the summer season June to September. A chi-square test of independence was performed between ISO method for E. coli and the bioanalyzer and between ISO method for E. faecalis and the bioanalyzer, p = 0.488 and p = 0.269, respectively. The novelty of the proposed method is that it integrates automated boiling as an extraction method to concentrate bacteria onto a filter, and it uses LAMP PCR to produce a portable optical sensor for environmental pathogen monitoring. This method is considered to be appropriate for bacterial detection in seawater, capable of monitoring coastal sites with results comparative to the gold standard method.
Keywords: biosensor; E. coli; E. faecalis; automated system; LAMP
Introduction
Approximately 41.2% of the European union population live in coastal areas and more than 50% of the global population live near a coastal area (Maul & Duedall, 2021) [1]. Coastal tourism accounts for 60% of all European tourists that prefer to spend their holidays at the seaside (cbi.eu). During 2017, 22.000 beaches and internal waters in Europe were tested for fecal indicator bacteria (FIB) and about 15% of the samples did not respond to “excellent” quality standard (EEA Report No 2/2018, 2018) [2]. 61 However, numerous human activities such as sewage effluents, agricultural inputs, malfunctions and random incidents from municipal sewage sources provoke microbial contamination to coastal waters and lead to public health risks. Despite the health promoting benefits derived from the use of bathing water, including health and wellbeing, social and cultural benefits, and economic value, users of bathing water may be exposed to a variety of aquatic organisms of public health concern, as microorganisms that may be harmful to or cause sickness in humans (Farrell et al., 2021; Sagarduy et al., 2019) [3,4].
Thus, microbiological indicators are used to assess the bathing water conditions. In European union, Water Framework Directive (WFD) and Marine Strategy Framework Directive (MSFD) establish a legal framework which guarantees the availability sufficient quantities of water of satisfactory quality throughout Europe (European Commission, 2006, 2008) [5,6]. The sanitary inspections consist of an on-field monitoring and evaluation of any visible features or conditions at bathing waters that may unbalance the quality of the seawater. Fecal exposure is evaluated through monitoring of two fecal indicator bacteria (Escherichia coli and intestinal enterococci), for mitigating hazardous dangers that can be related to public contamination to water polluted with feces. E. coli and enterococcus have been assessed as capable indicators for pathogenic waterborne bacteria because of their prevalence in the population and environment.
Thus, the European Directive recommends 250 CFU and 500 CFU of E. coli in 100 mL and as a “good quality” and “sufficient”, respectively. The limits for E. faecalis are formed to 100 CFU and 200 CFU in a seawater sample of 100 mL with the same characterization, as above (European Commission, 2006) [5]. During bathing season, at 84 least four samples require to be collected in each site. Water quality is categorized into four sections: poor, sufficient, good, and excellent. For excellent and good quality, the FIB must follow the predefined standards in 95% of samples. In case of sufficient classification, they must fulfill the requirements in 90% of samples.
For routine analysis, cultural classic methods are usually used as the method for counting FIB in seawater samples (European Commission, 2006) [5]. These methods are preferred because of easiness in operation, highly consistent and low cost (Tiwari et 91 al., 2016, 2018; Zhang et al., 2022) [7,8,9]. By applying these protocols, viable and culturable cells of FIB can be counted in order to correlate them with health risks factors (Bisimwa et al., 2022; Valenca et al., 2022) [10, 11]. Cultural methods need time for 94 sampling analysis and require at least 24-48 hours to export results because of the necessity for bacterial growth (Foddai & Grant, 2020) [12]. However, other studies dedicate on the identification of bacteria that cause fecal contamination through the use of methods involving biomarker enzymes which require dilution and an incubation step (Briciu-Burghina et al., 2019) [13]. Parallel to this, there are studies that attempt to infer indirectly the presence of fibroids by detecting metabolites that indicate fecal contamination (Zúñiga et al., 2022) [14]. Important plasmonic properties are present in nanomaterials. Thus, they are preferred for detection of trace pollutants in water because of their selectivity and sensitivity (Çimen et al., 2022) [15].
Molecular methods overcome some of the limitations that are faced up by classical methods. Polymerase Chain Reaction (PCR) methods have higher specificity and can enumerate targets more rapidly than cultural classic methods. Despite the fact that PCR is characterized by accuracy and reliability, it is an expensive tool to identify microbes in early stage of infection (Jasim et al., 2019) [16]. PCR like Loop-mediated isothermal amplification (LAMP), it doesn’t require excessive equipment, but it only amplifies the gene between 60°C and 65°C. Multiplex LAMP permits the recognition of multiple genes simultaneously. The comparative benefit of this molecular test is the time saving and cost-effective analysis (Rainbow et al., 2020) [17].
The fecal presence in seawater raises public concerns for population. Thus, there is a demand for a rapid device that could give precise results in the field. Of note, last years, biosensors have been emerged and assessed as a powerful tool for diagnosis (Andryukov et al., 2020) [18]. They are considered to be appropriate because they can be used in portable or in situ autonomous systems. Their principle is that they use a biorecognition element such as tissue, antibody, or a whole microorganism that in contact with a transducer transmits the information into a signal (Kotsiri et al., 2022) [19]. Biosensors will guarantee consistent and precise microbial hazard control to evaluate seawater state.
In this paper, we describe the simultaneous detection of E. coli and E. faecalis using an optical biosensor that uses a cellulose acetate (CA) filter and DNA extracted through boiling. This system is capable of performing a routine operation and gives precise results in the field as an automated system. As far as we are aware, this is the first instance of successfully using a membrane filter and the boiling method in conjunction with multiplex colorimetric LAMP for the detection of FIB.
Materials and Methods
Sampling locations: The study area was a part of Patraikos Gulf. The wider area is selected as a residential recreation area for swimming, fishing, and other recreational activities. (Figure 1) presents a schematic description of the seawater collection. The marine coast included 9 stations along the coastline starting from the Patras biological treatment area, with a high bacterial load, to the Rio area. The researchers determined sampling points by taking into account factors such as the sample’s proximity to the analysis area for instant analysis, secure access for data collection, the existence of a library from earlier analyses that were carried out in the laboratory and included the presence or absence of microorganisms in the sample, and swimming activities that took place both during the winter and during the summer, from June to September (Table 1). From each site, three samples were collected. Samples for every site were carefully collected in the morning, before a mass was held on the beach.
Figure 1: Map with the sampling stations in the area of Patras where water collection was carried out and analyzed with the classical laboratory methods and with the biosensor method.

Analyzer module: The individual parts that make up the final products are imported from overseas markets. The analyzer consists of three main parts: the sampling tank (effective volume: 1000 ml) and water filtration, the DNA extraction via boiling method and the detection phase. The system includes 8 2-way valves, two reservoirs (water sample, and backwash with sterile water), three peristaltic pumps, a peristaltic Peltier pump, three pumps, an air pump, a 3D printer (Creality Cloud), an electronic pipette variable volume 1-20 μL (0.1μL subdivision (Socorex)) (Figure 2), a microcontroller and a thermistor to control temperature. Microchips were created to facilitate the reading of the results in designated wells and easily visualize them. They consisted of two chambers in a slide format and were in-house fabricated by injection molding SYLGARD™ Silicone Elastomer (Dow Corning, USA). Each chamber had dimensions of R= 2.85 mm, corresponding to a volume of 18 μL. A 2MP Omnivision OV2640 was utilized for the viewing, supported by the Arduino programming environment, which was employed to categorize the data based on color changes.
Function of Bioanalyzer
(Figure 2) displays a schematic layout of the water sampling apparatus that is used to filter a sample of seawater and concentrate microorganisms. (Figure 3) shows an actual photo that was taken after construction was completed. The system includes two peristaltic pumps for the entrance of seawater into the 1 L tank and one pump for filter washing. The filtration of seawater is achieved with 70 (F1: Filter 1), 10 (F2: Filter 2) and 5 (F3: Filter 3) μm pore diameter membranes with a diameter of 50 mm and a height of 122 mm taking care to remove macro particles such as sand, plankton, parts of larger organisms (Figure 3). The 70 μm filter is made of stainless steel and it contains a washable pleated sleeve. The 10 and 5 μm filters are reusable and are made of woven polypropylene fibers. The inner ring is made of polypropylene and is ideal for removing suspensions, such as soil. The tank delivers one liter of seawater through a 0.45 μm CA-S syringe filter (Whatman, International Ltd.). CA filters used hydrophilic properties allowing reduced physisorption of organic material that could change the efficiency of backwash and resulted from the comparison of different types of membranes (Kotsiri & Vantarakis, 2023) [20]. After that, 100 ml of sterile water is used to wash the filter, and air is circulated to empty the tubing and eliminate any remaining water from the membrane surface. Then, the sample is transferred into a new tank and reverse washing is performed using 1 ml of sterile water to remove bacteria from the membrane. Iron surrounds the tank and is heated to break the bacteria’s cell wall (cube 1) (Figure 2).
Figure 3: Photo of model system after all modifications. A. three filters in a row B. air filter C. heating element D. cooling tanks E. touchscreen with system settings.

The next step involves transferring reagents, WarmStart® Colorimetric LAMP 2X Master Mix, primers mix E. coli and primers mix E. faecalis, (cubes 2,3,4) (Figure 2) via an electronic pipette. In this way, the simultaneous detection of microorganisms 183 in a seawater sample is achieved. Delivering the reagents together to mix them uniformly is done using a tank (cube 5). To clean the pipette from any leftover liquids it transported, another tank (cube 6) is utilized. Following agitation, the liquid is finally injected into a microchip well, where it is heated to 64°C for an hour to initiate the reaction.
The piping must be filled with seawater for the device to function properly. This also creates the necessary heating and cooling conditions for the reagents and for boiling the tank for DNA extraction. The peristaltic Peltier pump started to work and created a cooling and heating circuit. Water circulates in the pipes in order to secure temperatures in the system. The top side of the Peltier is in contact with the block containing the master mix and primers. Its underside is in contact with a cooling block, through which water flows to prevent the temperature from rising on the hot side of the Peltier. This keeps the reagents at -20°C while they are stored in the field and ensures that they are consistently at 4°C during the analysis.
Testing several seawater samples allowed for the evaluation of the bioanalyzer assembly.
LAMP reagents: The primers for the LAMP reactions, detecting E. coli and E. faecalis strains, were targeted to the region of the b-Dglucuronidase (uidA) (Ahmad et al., 2017) [21] and Enterococcus 23s rRNA genes (Lee et al., 2019a). Six primers total were used: two loop primers (LF and LB), two outer primers (F3 and B3) and two inner primers (FIP and BIP) (Eurofins, Germany).
The LAMP assay was performed in a total 20 μl mixture containing WarmStart® Colorimetric LAMP 2X Master Mix (New England Biolabs, Singapore), 10x LAMP primer mix (1.6 mM FIP and BIP, 0.2 mM F3 and B3, 0.4 mM of the LF and LB), 5.5 μL genomic DNA and sterile deionized water, according to the manufacturer’s instructions. The LAMP reactions were carried out at 64 °C for 60 min in triplicate.
Comparison with gold standard method: For the evaluation of the procedure, two protocols were applied to confirm the analyzer’s results, [23] (ISO 9308-1:2014) and [24] (ISO 7899-2:2014). The procedure was followed by passing 100 ml of seawater through a 0,45 um Mixed Cellulose Ester (Whatman, Maidstone, UK) membrane filter via vacuum pump. After filtration, the membrane was placed on CCA and SB plates, respectively. The dishes were incubated at 37°C for 16-18 hours for CCA plates and 48 hours SB plates. Then, the confirmation of the sample for E. faecalis, continued by incubation in Bile Aesculin Agar for two hours at 44°C. The results were assessed by calculating the number of 221 bacterial colonies on plates on a colony counter (CC-J3, Bioevopeak).
Statistical analysis: Statistical analyses were performed with the IBM SPSS Version 27.0 statistical software package. The association between ISO methods, for the detection of E. coli and E. faecalis, and bioanalyzer were studied using the chi-square test. A significance level was adopted (p<0.05).
Results
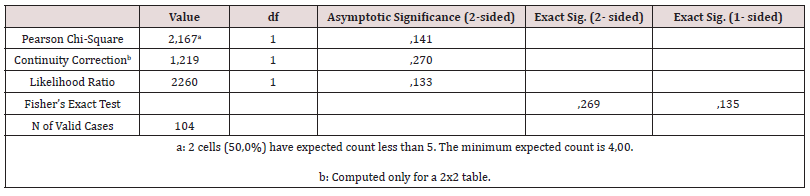
A total number of 52 seawater samples from different sites were collected. The colonies ranged between 0 and 1000 per 100 ml for the counting of E. coli and 0 to 227 per 100 ml for E. faecalis. The exact same samples were analyzed by bioanalyzer device. In the bioanalyzer system, the positive LAMP assay results were determined by the standard reagent’s, WarmStart Colorimetric, color change from pink to yellow. With the presence of DNA, ion transport is achieved and ultimately the visible color change in the sample with the reagent is succeeded. Actual photographs after microchip injection experiments and heating are shown in (Figures 4a&b). According to the analysis that performed with ISO method, the limit of detection of bioanalyzer is 5000 CFU/ 1000 ml seawater. The highest record of bacteria was 10000 CFU/ 1000 ml seawater. Thus, we increased the initial sample volume from 100 ml to 1000 ml in order to reach the desired detection limit. In a percentage of 94%, the measurements checked with the biosensor are identical to those that followed the traditional counting methods, comparing cultures in petri dishes with color changes after LAMP implementation. The chi-square test showed 0.488 for the comparison between ISO based method for E. coli with bioanalyzer and 0.269 ISO based method for E. faecalis with bioanalyzer.
Figure 4: Actual photos when extracting results. The negative sample is shown in bright pink and the positive in yellow.

These samples were boredly positive and were those ones that the seawater is characterized as sufficient.
A chi-square test of independence was performed to examine the relation between ISO method for E. coli and the bioanalyzer and between ISO method for E. faecalis and the bioanalyzer. The relation between these variables was not significant, X2 (1, N = 104) = 1.095, p = 0.488 and X2 (1, N = 104) = 2.167, p = 0.269, respectively (Tables 2 & 3). The two methods seem to give equal results.
Discussion
The reliability of the test was emerged from the comparison between bioanalyzer and gold standard method (ISO 7899-2, 2014; ISO 9308-1, 2014) [23,24]. Fifty-two samples of seawater were collected from nine sampling stations in the wider area of Patras. The stations were made up of coastal waters of various human activities, urban use (e.g. the port of Patras) but also swimming beaches and recreation (e.g. Rio beach). We further analyze the capacity of the biosensor system to correctly generate correct results by warning signal when water quality does not satisfy approved bathing thresholds. To evaluate the capacity of the suggested device to monitor seawater, we compared different sea samples. We have defined the threshold as the BWD suggests, upper than 500 CFU of E. coli in 100 mL and 200 CFU for E. faecalis in a seawater sample of 100 ml (European Commission, 2006) [5].
The difficulty of the analysis in seawater lies in the fact of the necessity for a preconcentration stage. In the laboratory, special equipment and the handling of the sample are required. In the field, techniques are needed to reduce the initial volume of the sample with methods. Specifically, in seawater samples a variety of methods has been used such as magnetic preconcentration using aptamers (Kotsiri et al., 2019; Pandit et al., 2022) [25,26], antibodies (Xiao et al., 2022) [27], filtration (Lee et al., 2019b) [28] and centrifugation (Wu et al., 2020) [29]. Laboratory methods are identical to the field application for using the filters in a device to achieve the preconcentration step, boiling method as an extraction technique has been assessed in different matrices (Huang et al., 2021; Wu et al., 2020) [29,30] and it has been suggested for bacterial lysis (Cao et al., 2022) [31]. The steps of the biosensor have been analyzed and evaluated as separate steps for their effectiveness (Kotsiri Z, Panteleli E, Vantarakis, 2022) [32]. The comparative advantage of the method is the simultaneous detection of microorganisms in seawater. The presence of one or the other organism automatically prohibits use by bathers.
Bioanalyzer device is an effective tool for routine monitoring of beach, bathing coasts in environmental sides. Other works have been set for water evaluation for E. coli counting, and Alert device is capable to monitor water samples after incubation (Angelescu et al., 2018; Briciu-Burghina et al., 2019) [13,33]. The performance of seawater analyzer device was confirmed by samples inoculated in the laboratory and tested manually according to the work (Kotsiri & Vantarakis, 2023) [20]. Another study that evaluates water samples for FIB is promising but it hasn’t been constructed in an integrated system (Lee et al., 2019b) [28].
Limitations of the Study
This work presents some limitations that could, however, be explored further in future research. According to the directives of the European Union, the classification of a beach as a sea will have to reach 500 CFU/ 100 ml. The present work had a minimum reproducible detection limit of 103 CFU/ 100 ml. Additional research is needed to eliminate the detection limit of the developed method. Of course, our work could overcome the problem by increasing the initial volume that was filtered. The biosensor is quite expensive in terms of its production cost with the specified components. Its size and weight they characterize it as inconvenient to move. The structure of biosensors requires a variety of consumables including molecular reagents and mechanical equipment. If the device is in operation, there is a need to keep all the reagents in refrigerated condition. any other temperature range would not give reliable results. Consequently, provision was made for the existence of a cooled container in which the reagents are deposited and cooled by contact. This system requires a continuous circulation of sea water, in order for the pelts that exist for the change in temperature to hold the vessel at low temperatures.
Moreover, the device needs a specific energy supply. The machine was built in such a way that it can be used battery as source supply. When the biosensor is to perform field analyzes and to pump water from the sea, a long water pipe is required which will transport the water to the device. Unfortunately, all the coasts don’t have the same environmental space so that it is possible to provide electricity based on the proximity to the biosensor that would be on the coast. Therefore, for this use, large batteries and panels are required to replace the current. The biosensors needed a specific current voltage both for their correct operation and to avoid damaged components due to overcharging.
As far as the engineering part is concerned, some difficulties were observed in the connection between the individual stages in the construction of a holistic system. For example, the material media carried fragments of the materials with the result that the water was not allowed to pass through the filter.
Acknowledgments
The present research received fund from the Single State Action Aid for Research, Technological Development & Innovation “INVESTIGATE - CREATE -INNOVATE” project “SMART-SEATRAC (T1EDK-04615)”
References
- Maul GA, Duedall IW (2021) Demography of Coastal Populations. In C. Finkl, C.W., Makowski (Ed.) Encyclopedia of Coastal Science. Encyclopedia of Earth 422 Sciences Series
- EEA Report No 2/2018 (2018) European bathing water quality in 2017. European Environment Agengy Reports 2: 1-28.
- Farrell ML, Joyce A, Duane S, Fitzhenry K, and Hooban B, et al., (2021) Evaluating the potential for exposure to organisms of public health concern in naturally occurring bathing waters in Europe: A scoping review. Water Research 206(September).
- Sagarduy M, Courtois S, Del Campo A, Garmendia JM, and Petrau A, et al., (2019) Differential decay and prediction of persistence of Enterococcus spp. and Escherichia coli culturable cells and molecular markers in freshwater and seawater environments. International Journal of Hygiene and Environmental 437 Health 222(4): 695-704.
- European Commission (2006) Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 concerning the management of bathing water quality and repealing Directive 76/160/EEC. Official Journal of the European Union L 064(1882): 37-51.
- European Commission (2008) Directive 2008/56/EC of the European Parliament and of the Council of 17 June 2008, establishing a framework for community action in the field of marine environmental policy (Marine Strategy Framework 372 Directive). Journal of Water Law 19(3): 95-97.
- Tiwari A, Niemelä SI, Vepsäläinen A, Rapala J, and Kalso S, et al., (2016) Comparison of Colilert-18 with miniaturised most probable number method for monitoring of Escherichia coli in bathing water. Journal of Water and Health 14(1): 121-131.
- Tiwari A, Hokajärvi AM, Santo Domingo JW, Kauppinen A, and Elk M, et al., (2018) Categorical performance characteristics of method ISO 7899-2 and indicator value of intestinal enterococci for bathing water quality monitoring. Journal of Water and Health 16(5): 711-723.
- Zhang J, Li C, Rahaman MM, Yao Y, and Ma P, et al., (2022) A comprehensive review of image analysis methods for microorganism counting: from classical image processing to deep learning approaches. In Artificial Intelligence Review 55(4): 2875-2944.
- Bisimwa AM, Kisuya B, Kazadi ZM, Muhaya BB, and Kankonda AB, et al., (2022) Monitoring faecal contamination and relationship of physicochemical variables with faecal indicator bacteria numbers in Bukavu surface waters, tributaries of Lake Kivu in Democratic Republic of Congo. Hygiene and 347 Environmental Health Advances 3(June) 100012.
- Valenca R, Garcia L, Espinosa C, Flor D, and Mohanty SK, et al., (2022) Can water composition and weather factors predict fecal indicator bacteria removal in retention ponds in variable weather conditions? Science of the Total Environment 838(February): 156410.
- Foddai ACG, Grant IR (2020) Methods for detection of viable foodborne pathogens: current state-of-art and future prospects. Applied Microbiology and Biotechnology 104(10): 4281-4288.
- Briciu-Burghina C, Heery B, Duffy G, Brabazon D, and Regan F, et al., (2019) Demonstration of an optical biosensor for the detection of faecal indicator bacteria in freshwater and coastal bathing areas. Analytical and Bioanalytical Chemistry 411(29): 7637-7643.
- Zúñiga A, Muñoz-Guamuro G, Boivineau L, Mayonove P, and Conejero I, et al., (2022) A rapid and standardized workflow for functional assessment of bacterial biosensors in fecal samples. Frontiers in Bioengineering and Biotechnology 10(August).
- Çimen D, Bereli N, Denizli A (2022) Advanced Plasmonic Nanosensors for Monitoring of Environmental Pollutants. Current Analytical Chemistry 19(1): 2-17.
- Jasim I, Shen Z, Mlaji Z, Yuksek NS, and Abdullah, et al., (2019) An impedance biosensor for simultaneous detection of low concentration of Salmonella serogroups in poultry and fresh produce samples. Biosensors and Bioelectronics 126: 292-300.
- Rainbow J, Sedlackova E, Jiang S, Maxted G, and Moschou D, et al., (2020) Integrated electrochemical biosensors for detection of waterborne pathogens in low-resource settings. Biosensors 10(4): 36.
- Andryukov BG, Besednova NN, Romashko RV, Zaporozhets TS, and Efimov TA, et al., (2020) Label-free biosensors for laboratory-based diagnostics of infections: Current achievements and new trends. Biosensors 10(2): 11.
- Kotsiri Z, Panteleli E, Vantarakis A (2022) Evaluation of Molecular Methods to Be Used in Sensors for The Detection of Bacteria in Water. Journal of Biosensors & Renewable Sources 235-242.
- Kotsiri Z, Vantarakis A (2023) Evaluation of seawater monitoring for the detection of Escherichia coli and Enterococcus faecalis on an integrated biosensor system. 43: 99-98(7).
- Ahmad F, Stedtfeld RD, Waseem H, Williams MR and Cupples AM, et al., (2017) Most probable number - loop mediated isothermal amplification (MPN-LAMP) for quantifying waterborne pathogens in < 25 min. Journal of Microbiological Methods 132: 27-33.
- Lee S, Khoo VSL, Medriano CAD, Lee T, and Park SY, et al., (2019a) Rapid and in-situ detection of fecal indicator bacteria in water using simple DNA extraction and portable loop-mediated isothermal amplification (LAMP)PCR methods. Water Research 160: 371-379.
- ISO 9308-1 (2014) ISO 9308-12014AMD 12016.
- ISO 7899-2 (2014) ISO 7899-2_2014(en), Water quality -Detection and enumeration of intestinal enterococci-Part 2_ Membrane filtration method. ISO 7899-2_2014, 2000, 7899.
- Kotsiri Z, Vantarakis A, Rizzotto F, Kavanaugh D, and Ramarao N, et al., (2019) Sensitive detection of E. coli in artificial seawater by aptamer-coated magnetic beads and direct PCR. Applied Sciences 9(24).
- Pandit C, Alajangi HK, Singh J, Khajuria A, and Sharma A, et al., (2022) Development of magnetic nanoparticle assisted aptamer-quantum dot-based biosensor for the detection of Escherichia coli in water samples. Science of the Total Environment 831(December 2021): 154857.
- Xiao S, Yang X, Wu J, Liu Q, and Li D, et al., (2022) Reusable electrochemical biosensing platform based on egg yolk antibody-labeled magnetic covalent organic framework for on-site detection of Escherichia coli in foods. Sensors and Actuators B: Chemical 369 (February) 32320.
- Lee S, Khoo VSL, Medriano CAD, LeeT, and Park SY, et al., (2019b) Rapid and in-situ detection of fecal indicator bacteria in water using simple DNA extraction and portable loop-mediated isothermal amplification (LAMP)PCR methods. Water Research, 160: 371-379.
- Wu X, Huang X, Zhu Y, Li J, and Hoffmann MR, et al., (2020) Synthesis and application of superabsorbent polymer microspheres for rapid concentration and quantification of microbial pathogens in ambient water. Separation and Purification Technology 239:116540.
- Huang C, Mahboubat BY, Ding Y, Yang Q, and Wang J, et al., (2021) Development of a rapid Salmonella detection method via phage conjugated magnetic bead separation coupled with real-time PCR quantification. Lwt 142: 111075.
- Cao Y, Ye C, Zhang C, Zhang G, and Hu H, et al., (2022) Simultaneous detection of multiple foodborne bacteria by loop mediated isothermal amplification on a microfluidic chip through colorimetric and fluorescent assay. Food Control 134: 108694.
- Kotsiri Z, Vidic J, Vantarakis A (2022) Applications of biosensors for bacteria and virus detection in food and water-A systematic review. Journal of Environmental Sciences (China) 111: 367-379.
- Angelescu DE, Huynh V, Hausot A, Yalkin G, and Plet V, et al., (2018) Autonomous system for rapid field quantification of Escherichia coli in surface waters. Journal of Applied Microbiology 126(1).

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...