Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2641-1687
Research Article(ISSN: 2641-1687) 
Preparation of Doxorubicin-Retinoic Acid Hip Complex Nanomicellar Formulation for Doxorubicin Delivery to Prostate Cancer
Volume 1 - Issue 4Alex Oselu Owiti1*, Dhananjay Pal2 and Ashim Mitra2
-
Author Information
Open or Close
- Department of Pharmaceutical Sciences, School of Pharmacy, University of Missouri-Kansas City, USA
- Department of Pharmaceutical sciences, University of Missouri Kansas City.
*Corresponding author:
Alex Oselu Owiti PhD, Department of Pharmaceutical Sciences, School of Pharmacy, University of Missouri- Kansas City, USA
Received: February 26, 2019; Published: March 07, 2019
DOI: 10.32474/JUNS.2019.01.000118
Abstract
Doxorubicin is a hydrophilic anticancer drug. Due to hydrophilicity, it’s difficult to encapsulate in a hydrophobic core of nanomicelles. The main purpose of this study was to develop a hydrophobic ion paring complex (HIP) of doxorubicin using hydrophobic retinoic acid (Vitamin A). The resultant hydrophobic (DOX-RA) complex was utilized to prepare drug-loaded nanomicelles by co-precipitation method with penta block copolymer. Nanomicelles (DOX-RA/PBNM), was prepared by evaporation rehydration technique. DOX-RA complex was analyzed by H-NMR and FTIR. The nanomicelles were analyzed for size and zeta potential using dynamic light scattering (DLS) as well as transmission electron microscopy (TEM). The FT-IR and the H-NMR analyses confirmed the structures of the DOX-RA complex. Mean nanomicellar sizes were 25.5nm ± 5.00nm, and ζ-potential was approximately zero. H-NMR and FTIR analysis done on DOX-RA indicate peaks characteristic of both doxorubicin and retinoic Acid. This confirmed successful complex formation. Transmission electron microscopy (TEM) analysis revealed round shaped morphology and sizes similar to DLS results. In vitro release studies revealed that pentablock nanomicelles released Doxorubicin at a slow first order rate in phosphate buffer solution (PBS) at pH 7.4 compared to pH 5.5 and pH 4.0. Confocal microscopy analysis with DOX-RA/ PBNM indicated that pentablock nanomicelles were efficiently taken into prostate cancer (PC3) cells and doxorubicin was efficiently released from the nanomicelles into the cells. In addition, cell proliferation assay showed that nanomicelles ferried adequate amounts of Doxorubicin into PC-3 cells and inhibited cell growth significantly. Results confirm that DOX-RA complex facilitated the encapsulation of doxorubicin within nanomicelles increasing DOX intracellular concentration.
Introduction
Doxorubicin (DOX), available in the market as Adriamycin or Rubex, is a drug used in treatment of various cancers. Doxorubicin is a member of anthocyanins family and is grouped under anthracycline with both antitumor and antibiotic activities. It works by interfering with DNA function causing intercalation and alkylation of DNA, causing disruption to both RNA and DNA polymerase, inhibition of topoisomerase II which consequently kills the tumor cells [1,2]. DOX is applied mostly in the treatment of breast and bladder cancers. It is also used in cases of sarcomas, lymphomas, and acute lymphatic leukemias. It is normally used as combination with other drugs by intra-venous application. However, there are many serious side effects associated with doxorubicin [3]. These side effects include but not limited to allergic reactions which may cause tissue damage at the spot of injection. Another most notorious side effect is the accumulation of doxorubicin in the cardiac and respiratory tissues. This irreversible dose-dependent, side effect may cause toxicity to cardiac and respiratory tissues, resulting in cardiomyopathy, dyspnea and intolerance to exercise due to the production of mitochondrial reactive oxygen species (ROS). Other common collateral effects are loss of hair, suppression of bone marrow, skin eruptions, vomiting and mouth inflammation. Patients often experience red coloration of the urine for a few days during treatment with DOX. Doxorubicin is a widely used drug in clinical setting. However, less entry and low distribution of doxorubicin in tumor tissue are the principal factors for its therapeutic backlash [3]. Due to low pH in the tumor interstitial environment, weak base drugs like doxorubicin, are likely to ionized before entering leading to reduced cell uptake [4].
In addition, serious cytotoxic effects may be caused to healthy cells due to non-targeted delivery. These may result in dose-dependent cardiac damages, multidrug resistance, and myelosuppression thus, restricting its therapeutic application [5]. It is important to develop new delivery systems that capable of ferrying sufficient amount of drug to the tumor cells, thereby avoiding the multidrug resistance and frequent dose administrations of chemotherapeutics. One of the attempts to elevate the drug bioavailability in tumor tissue is the application of site-specific delivery systems that may release the encapsulated drugs within tumor cells. Another way is to attain higher accumulation of drug by specific tumor cell targeting. However, this may be difficult to achieve without using a ligand that is unique to the target on the cell surface. This lack of proper ligand may end up delivering the drug to the undesired site [6]. Some intrinsic variations in the tumor microenvironment such as enzymes, pH and oxidative stress, and the extrinsic factors i.e., light, temperature and magnetic fields, are known to cause site-specific drug release within the tumor cells and tissues [7]. pH triggered drug release from a nanocarrier is the most acceptable way of drug release directly inside the cell cytoplasm [7]. This strategy of using the acidic tumor microenvironment to trigger drug release has shown some short falls due to the fact that the lowest pH a tumor is located far from tumor blood vessels [8]. Since the interstitial region of a tumor has a pH lower than 6.5, the ability nanocarriers might be restricted. For instance, some carriers such as liposomes become unstable and are altered before arriving at their intended targets. Another promising type of nanoformulation for delivery of hydrophobic drugs is nanomicelles. Nanomicelles display higher efficiency compared to other carriers like liposomes.
They also possess many advantages, which include high bioavailability, improved stability of the encapsulated drug, better encapsulation and loading efficiencies and better delayed release profile [9]. Furthermore, most of the materials utilized in its production are biologically safe for all forms of administration. The application of these nanomicelles is with respect to ability to improve drug entry into the tumor tissue, while reducing dosage and enhancing drug efficacy by limiting non-selective cytotoxicity [10-13]. To cause pH-triggered intracellular release of doxorubicin (DOX), retinoic acid (RA), a lipophilic molecule, was utilized to form an ion paired complex (DOX-RA). The formed ion pair shows instability in an acidic microenvironment i.e. pH < 7.0, which is compatible with tumor tissue. The complex attains stability in an environment with pH 7.4, for instance, blood [14]. This would help in increasing the drug levels in the cells of tumor tissue, sparing normal tissues cells. In addition, hydrophobicity caused by ion pairing can retain doxorubicin in the hydrophobic core of nanomicelles and raise the encapsulated amount of DOX in the nanomicelles. Furthermore, studies have revealed that RA may improve effectiveness of a drug by interfering with the permeability and fluidity of membrane of tumor cells [15].
Materials and Methods
Retinoic acid was purchased from Aeser, MA, USA. Pentablock copolymer containing Poly-ethylene glycol (PEG), L-lactide and ε-caprolactone monomers, was obtained from our laboratory, already synthesized from previous project. CyquantTM cell proliferation assay kit was obtained from Invitrogen Life Technologies Inc. and distributed by Thermos-Fisher Scientific. Doxorubicin drug was obtained from ADOOQ Bioscience, Irvin, CA, USA. All other reagents utilized in this study were of analytical grade.
Preparation of DOX-RA
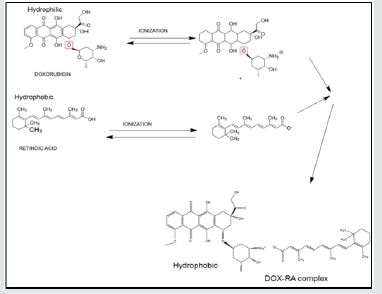
In this study, a hydrophobic complex of doxorubicin and retinoic acid (DOX-RA) was prepared by co-precipitation technique. Briefly, doxorubicin aqueous solution (10 mg/mL), was prepared in an Eppendorf tube, 100mg/mL sodium bicarbonate solution was added to the tube while stirring. An ethanolic solution of retinoic acid (10 mg/mL), was added to the DOX solution while stirring continuously. After stirring the combined solutions for 1 h, the final solution was centrifuged at 6000 rpm for 20 min. Finally, an orange to brick-red colored precipitate of DOX-RA (Figure 1) settled at the bottom of the tube. The precipitated complex was washed 3 times with deionized water, followed by drying at 40 °C before being stored at 4°C. The DOX-RA complex was then used to prepare nanomicelles with pentablock copolymer previously prepared in our lab [16].
Figure 1: The schematic illustration of the reaction and the structures of DOX, RA and DOX-RA complex.
Determination of Hydrophobicity DOX-RA Complex
Hydrophobicity of DOX-RA complex was analyzed by studying its solubilities in octanol. DOX-RA was dissolved in octanol, while agitating for 48h at room temperature (25°C). The sample was then submitted to centrifugation for 20min at 6000 rpm. After centrifugation, the supernatant was collected and dissolved in isopropanol, homogenized and then analyzed using UV-vis spectrophotometer (Shimadzu, Kyoto, Japan) 495nm wavelength.
FTIR Analysis of DOX-RA Complex
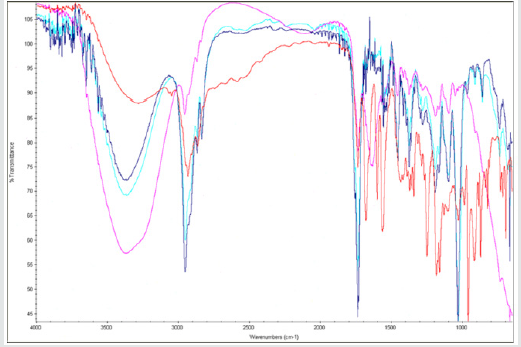
To determine structural characteristic of the formed, DOX-RA complex, and the success of the complex formation. Samples of synthesized DOX-RA complex, RA and DOX were subjected to FTIR, Thermo-Scientific Nicolet iZ10, with an ATR diamond and DTGS detector. Where samples of DOX-RA complex, DOX and RA were analyzed at scanning range of 650–4000 cm−1.
Mass Spectrometry Analysis for DOX-RA complex
The DOX-RA complex was submitted to mass spectrometry (Thermo LCQ deca mass spectrometer). The analysis with LC-MS/ MS involved a positive ion mode with electrospray ionization (ESI) as the main ion source. The source was set at 5 kV, the rates of both sheath gas and auxiliary gas flow were set at 80 and 20 units, respectively while capillary temperature was adjusted to 250 °C. C-18 column of 2.0mm × 50mm, and particle size limit 3 μm was used for Liquid Chromatography separation applying using water with 0.1% formic acid as the mobile phase A and acetonitrile as the mobile phase B. The flow rate of LC was adjusted to 0.20 ml/min. The gradient of mobile phase B increased from 30% to 100%.
Preparation of DOX-RA Nanomicelles
Nanomicelles(NM) were prepared using hydration-dehydration method [17]. Briefly, a specific amount (0.1%w/v) of DOX-RA complex was dissolved in 1ml of ethanol in an Eppendorf tube. On the other hand, 2%w/v of pentablock copolymer was dissolve in 1 mL ethanol in a separate tube. The two solutions were then mixed together, homogenized and evaporated in a speed vacuum for 24h until the formation of a thin film. Resultant thin film was suspended in deionized (DI) water followed by vortexing until a complete dissolution was achieved [18]. The solution was filtered using 0.22μm filter membrane to acquire uniform nanomicelle sizes and eliminate the polymer residues. The nanomicelle suspension was further freeze-dried using LABCONCO freeze-dryer, USA, with trehalose as cryo-protectants [2].
Determination of Nanomicelle Size and Zeta Potential
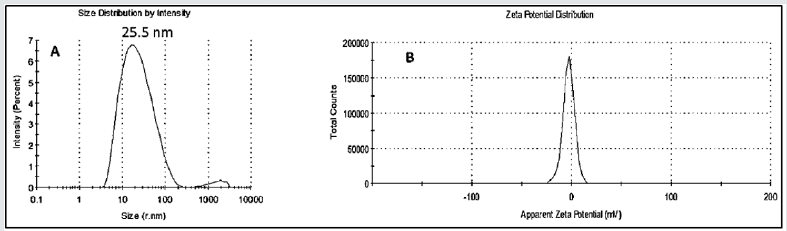
The sizes, zeta potentials and polydispersity indices (PDI) of DOX-RA-PBNM were obtained using dynamic laser scattering (DLS) with the help of Zetasizer HS 3000 (Malvern Instruments, UK), at a detection angle of 90 degrees at 25oC. The samples were dissolved in deionized water, homogenized and filtered using 0.22μL before being transferred into a transparent cuvette for analysis. Average values of three measurements were obtained for all samples and the peaks for zeta sizes, PDI and zeta potentials were obtained and recorded accordingly [19].
Determination of Drug Encapsulation and Loading Efficiencies
The amount of doxorubicin encapsulated in the nanomicelles was measured by UV-vis. briefly, a portion of the nanomicellar formulation was freeze-dried. Then 2mg of the dry formulation was dissolved in 1mL dichloromethane (DCM) to break the nanomicelle structure followed by centrifugation at 10,000rpm for 5min and then eventual evaporation to remove DCM. Subsequently, dry content was collected and dissolved in ethanol. The resultant ethanolic solution was then filtered through 0.22-mm nylon syringe filter. The filtrate was analyzed with UV-spectrophotometry at 495nm (Beckman, DU® 530, UV-vis spectrophotometer, Life science, CA, USA), for the determination of doxorubicin concentration. This process was performed in triplicate and a calibration curve with pure doxorubicin dissolved in ethanol at different concentrations produced an equation Y=42.024+0.031 and R2 =0.9884 [20]. Percentage encapsulation efficiency (%EE) and drug loading were calculated according to Eqs. 1 and 2.
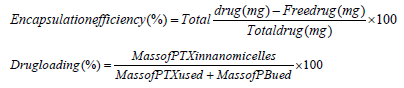
H1-NMR Analysis for DOX-RA-PBNM
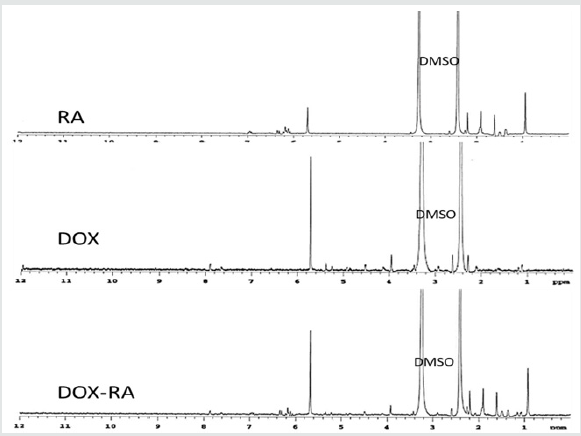
DOX-RA-PBNM were analyzed using deuterated dimethyl sulfoxide (DMSO) as solvent then subjected to H1-NMR analysis. An adequate amount of both pure doxorubicin and retinoic acid were dissolved in DMSO before analysis. The spectra were obtained at room temperature upon submission to a Varian Inova 400 MHz spectrometer (Varian, Palo Alto, CA, USA). Chemical shifts values were reported in parts per million (ppm) [21]. 1H-NMR spectra were recorded from 0 to 12 ppm using a delay time of 4 seconds.
Transmission Electron Microscopy Analysis
Nanomicelles morphology was determined by transmission electron microscopy (TEM; Philips CM12 STEM, Hillsboro, OR.). Briefly; a small amount of freeze-dried DOX-RA-loaded pentablock nanomicellar formulations were dissolved in DI water to form aqueous solution. The samples were then stained with 1% uranium salt. The resultant solution containing nanomicelles was then placed on a carbon-coated copper grid, and excess liquid was removed using a piece of dry filter paper then submitted to transmission electron microscope.
In vitro Release Study for DOX-RA Nanomicelles
The release of doxorubicin from DOX-RA/PBNM was conducted in various media of different pH conditions (pH 4.0, pH5.5 and pH 7.4) for 108 h in a dark room [22]. Briefly, 2mL of (2%w/v), DOX-RA/PBNM aqueous solution was transferred into a dialysis membrane bag (MW 10kDa) [23]. The dialysis bags were dipped into 15 mL tubes of release media and placed in a shaking water bath at 37 °C. The media were completely replaced at different time intervals i.e. 3 h, 6 h, 12 h, 24 h, 36 h,48 h, 60 h, 72 h,84 h,96 h and 108 h respectively [24]. The doxorubicin amount released from DOX-RA-PBNM was obtained with a UV spectrophotometer using 495nm wavelength [25].
Cell Uptake Analysis of DOX-RA-PBNM
During the uptake studies, confocal laser scan microscopy was applied to determine the intracellular distribution of both DOXRA/ PBNM in PC-3 cells. PC-3 cells were cultured until confluence in two different 4 chamber slides using appropriate growth media. A solution of DOX-RA-PBNM 20 μg/mL was made with culture medium. The solution was transferred into each 4 chamber slides containing PC-3 cells and incubated for different time intervals i.e., one was incubated for 12 h and the other for 24 h. The slides were treated with 4%paraformaldehyde, washed 3 times with PBS, and then kept in mounting medium overnight. The slides were then observed using confocal scanning microscopy (Leica microsystems, wezler, Germany) at 340nm and 540 excitation and emission wavelengths respectively [21,26].
In vitro Growth Inhibition Assay for DOX-RA-PBNM
Cell growth inhibition analysis was conducted using a DNA based Assay Kit called CyquantTM® Cell Proliferation (C7026) that contains fluorescent dye called CyquantTM® GR dye [27]. Briefly; PC-3 cells were cultured in a 96 well plate with Dulbecco’s F-12 medium as per protocol. Cells were incubated in 96 well plate at 37 OC and 5%CO2 until confluence. The medium was then removed followed by addition of different medium solutions made of pure doxorubicin (20μg/mL) and DOX-RA/PBNM (20μg/mL) in specific rows of the plate. The plate was then incubated under humidified atmosphere at 37°C and 5% CO2 for 24 h and 48 h. in each case, Cyquant reagent solution was prepared and added in each well after washing 3 times with PBS. The plate was kept at 4oC before being submitted to fluorescent plate reader. The readings were recorded at wavelengths of 570 and 630 nm. All the measurements were done in triplicates and reported as mean plus or minus standard deviation.
Results and Discussions
Hydrophobicity Analysis for DOX-RA Complex
Hydrophobicity of DOX-RA complex was analyzed before the preparation of DOX-RA nanomicelles. The lipid solubility of saturated DOX-RA was measured in octanol in order to assess the impact of ion pairing on lipophilicity of doxorubicin [28]. The solubility of doxorubicin in hydrophobic media improved significantly after the formation of ion pair with RA, which is a clear indicator of a high hydrophobicity caused by a successful complexing of DOX with RA. The enhanced lipophilicity of doxorubicin with retinoic acid ameliorate drug loading and encapsulation of DOX inside PBNM.
Mass Spectrometry Analysis for DOX-RA Complex
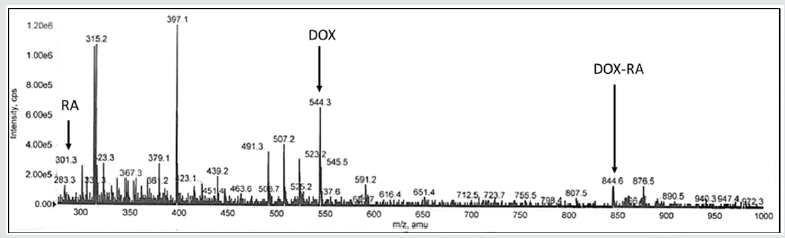
Mass spectrometry was conducted to confirm the constitution of the DOX-RA complex with the above-mentioned MS/MS conditions. The ion peaks were observed for both DOX and RA at different m/z. In positive ion mode MS/MS analysis, major fragment peaks of both RA and DOX ([M+H]+) were observed at m/z 301.3 and 544.3 respectively, while molecular ion peaks at m/z 844.6 were observed for the DOX-RA complex (Figure 2), this being the sum of both RA and DOX molecular weights(DOX=544g and RA=300g). The result shows evidence that DOX-RA complex was constituted of both doxorubicin (Mw 544) and Retinoic acid (Mw 300g).
Figure 2: The mass spectrometry spectra showing the Mz peak for RA, DOX and DOX-RA complex in a positive ion mode.
FTIR Analysis of DOX-RA Complex
FTIR spectra were obtained for dry samples of DOX-RA as well as pure doxorubicin drug and Retinoic acid samples. The results showed increase in absorption band at 1700 cm−1 and multiple bands that range from 1000–1500 cm−1 depicting the presence of increase in all the significant stretching’s bands in DOX-RA as a result of complex formation. This is in comparison to the individual molecules of DOX and RA. As shown in Figure 3. C=O bands at 1050 cm−1 and O-H band stretching at 3300 cm−1 are for DOX and RA. C-H stretching bands at 2940 and 2900 cm−1 indicate the presence of both DOX and RA. Bands manifested in the region around 3000cm -1 can be assigned to aromatic C-H stretching of both DOX and RA in the DOX-RA complex. There is an increase in C-C ring stretching peak in DOX-RA complex compared to the individual molecules of both DOX and RA in the region around 1750cm-1. However, primary aliphatic amines in DOX is constant in the region 3450-3250 cm-1.
1H-NMR Analysis for DOX-RA Complex
H-NMR analysis was conducted in order to understand the structure of the newly formed DOX-RA. DMSO was used as solvent to analyze the samples. DOX-RA/PBNM, free DOX and Free RA were dissolved in DMSO. Although DOX is hydrophilic, DMSO was used for all the samples due to its ability to dissolve both hydrophobic and hydrophilic substances. Samples containing DOX-RA complex in DMSO showed sharp peaks representing various protons of DOX and RA. Interestingly, a sample containing DOX-RA/PBNM in DMSO was devoid of any signal of signal of the substances suggesting that DOX-RA is inside the nanomicellar core. In the nanomicelle sample, broad peaks observed at 3.75 δ ppm and 2.30 δ ppm were attributed to the protons of DOX (-CH2-CH2-) and (-CH3) of RA, respectively Figure 4. Proton peaks for the DOX and RA in the complex were very eminent. These results suggest that DOX-RA complex was successfully formed.
Size Distribution and Zeta Potential for DOX-RA Loaded Nanomicelles
Solvent evaporation method was applied for the preparation of DOX-RA/PBNM. The PBNM formulations were analyzed to determine the size, polydispersity index (PDI), and zeta-potential. Average mean diameters for the nanomicelles (DOX-RA/PBNM) was 25.5±5.00nm (Figures 3 & 4). Small negligible peaks appeared at regions beyond 1000nm for DOX-RA/PBNM, perhaps due to a small aggregation in the sample, but still the majority peak (around 98%) was 25.5±5.00nm (Figure 5A). Average PDI for DOX-RA/ PBNM 0.037±0.005. PDI values suggest that the nanomicelles exhibit uniform particle size distribution with less aggregation. The drug-loaded nanomicelles display neutral zeta potential (around 0±2.5 mV) as shown in Figure 5B. This makes it possible for the DOX-RA nanomicelles to be an excellent candidate for intravenous application since it is unlikely to produce interactions with blood components due to neutrality.
Entrapment Efficiency and Drug Loading of DOX-RA Nanomicelles
Percentage of drug encapsulation efficiency (EE) is a crucial factor for drug delivery carriers. The percentage EE of doxorubicin loaded in DOX-RA nanomicelles was calculated to be 97.5% while the drug loading was found to be approximately 12%. These results demonstrate that DOX-RA pentablock nanomicelles achieved high doxorubicin entrapment efficiency and adequate drug loading which might be attributed to the hydrophobicity caused to DOX by complexing it through ion pairing with retinoic acid, thereby creating its efficient lodging in the hydrophobic core formed by hydrophobic PCL and PLA of the pentablock nanomicelle. This technique allows for efficient entrapment of doxorubicin drug in the nanomicellar core, which would have been hard to achieve due to hydrophilicity of doxorubicin. The encapsulation efficiency of the formed DOX-RA/PBNM was 98.5%, which is almost 100% of doxorubicin encapsulation. Furthermore, the percentage drug loading was 12%. This might be attributed to lipophilicity caused by ion pairing of DOX with RA. In addition, the lodging of high concentration of DOX-RA inside the hydrophobic core of PBNM shows the success in the complex formation and increased hydrophobicity of doxorubicin.
Analysis by Transmission Electron Microscopy (TEM)
Figure 6: The TEM image showing the DOX-RA-PBNM after freeze-drying using trehalose as cryoprotectant.
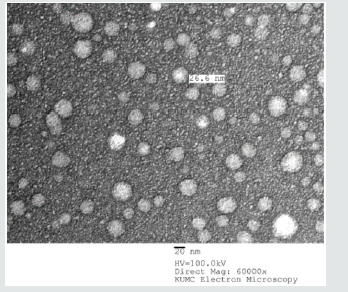
The morphology of pentablock nanomicelles containing DOXRA complex was investigated by transmission electron microscopy (TEM). This analysis showed that the nanomicelles were spherical, homogenous, and did not contain aggregates (Figure 6). Regarding size, pentablock nanomicelles had an average of 30±2.5nm. There was an apparent uniform morphology in DOX-RA/PBNM, with uniform appearance. The particle sizes visualized in the TEM images corroborate with the sizes obtained by DLS which already depicts the DOX-RA/PBNM and the empty PBNM sizes as 30nm ±5.00nm.
In vitro Release Study for DOX-RA-PBNM
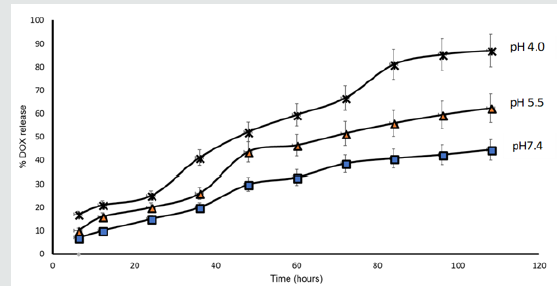
The drug release profile of doxorubicin from DOX-RA/ PBNM was analyzed for 108 h at 37 °C under different conditions i.e., pH 7.4 and pH 5.5 and pH4.0 phosphate buffer solutions. The acidic (pH 4.0) medium was applied to simulate the acidic microenvironment of cancer cells. The cumulative percentage release of DOX from DOX-RA/PBNM is shown in Figure 7 where pH-dependent doxorubicin release from DOX-RA Nanomicelles was observed. The DOX release at pH 7.4 occurred slowly in a sustained manner, with approximately 40% of DOX being released within 108 h. This indicates that if administered through IV, DOX-RA/PBNM could keep the drug in blood circulation for a long time. Studies have revealed that the amount of drug released from a carrier like nanomicelle is suppressed while in systemic circulation [29]. However, in this study, the release profile in low pH media (pH 5.5 and pH 4.0) occurred relatively faster with time compared to that in pH 7.4. There was high doxorubicin release (60% and 80%) after 108 h in media conditions of pH 5.5 and pH 4.0, respectively. This may be due to the formation of cations of doxorubicin at acidic pH, which increased its hydrophilicity thus returning it to its original form [4]. These results indicate that DOX-RA complex can keep a large amount of DOX within the core of nanomicelles for a long time in the blood stream upon intravenous administration. The drug release in the PC-3 cells may have been enhanced by the low pH within the lysosomes of tumor cells or due to the acidic microenvironment inside the cellular endosomes while the cell uptake of nanomicelles occurred by.
Figure 7: In vitro release profiles of DOX from DOX-RA/PBNM in pH 7.4, pH 5.5 and pH 4.0 media at 37 ± 0.5 °C. recorded as the mean ± SD (n = 3).
In vitro Cell uptake studies of DOX-RA/PBNM
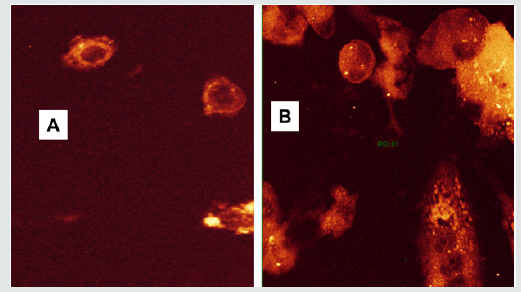
Cell uptake study was conducted to ascertain the presence of DOX inside the cells after exposure to the Nanomicelles. DOXRA loaded pentablock nanomicellar formulation was incubated with Prostate cancer PC-3 cells for 12 h and 24 h under conducive conditions of 5% carbon dioxide, 37o C (30). An appropriate media for the growth of the cells was used [30-32]. Cells were incubated in chamber slides with pentablock nanomicelles bearing DOX-RA complex [33,34]. The results of this study showed that there was a significant uptake of the nanomicelles into prostate cancer (PC- 3) cells, with higher amount of drug entering the cells after 24 h compared to 12 h as shown in Figure 8 [35-37]. In Figure 8B, brickred DOX was observed with high intensity all over the cytoplasmic region including the nucleus. Therefore, it can be concluded that fluorescent DOX molecules are inside the cells. This indicates that nanomicelles were internalized by PC-3 cells. This is an evidence that a large amount DOX was taken inside the cells with DOX-RAPBNM and the release was maximum afterv24 h (Figure 8B). The intensity of fluorescence is therefore directly proportional to the amount of DOX that enter the cells. Figure 8 shows a confocal image of PC-3 cells observed after incubation with DOX-RA/PBNM for 12 h and 24 h. The fluorescence intensity within cells treated with DOX-RA/PBNM for 24 h was higher than that of cells treated for 12 h. Fluorescence is very explicitly visible all over the cytoplasmic region after 24 h incubation. The cell uptake amount of DOXRA/ PBNM increased with time in PC-3 cells. After cell uptake, the nanomicelles could be degraded by lysosomal enzymes and other related factors within the cells, resulting in their structural disruption and subsequent release of doxorubicin DOX inside the cytoplasm. There was a significant amount of DOX-RA- loaded nanomicelles inside the cytoplasm (p < 0.05). This observation suggests that high uptake of the drug loaded DOX-RA-PBNM nanomicelles may have been due to endocytosis. The uptake and distribution of DOX-RA/PBNM by PC-3 cells that was observed by confocal microscopy could be related to the anti-tumor efficacy of DOX. Since DOX itself is fluorescent, it could be applied to directly measure cell uptake without applying any fluorescent marker.
Figure 8: Confocal images featuring the time dependent (A) 12 h and (B) 24 h) intracellular uptake of DOX-RA/PBNM by PC-3 cells.
In vitro Growth Inhibition Assay for DOX-RA-PBNM
Figure 9: The cell proliferation assay graphs showing %Cell survival after treatment of PC-3 cells with DOX-RA/PBNM, Free DOX for 24 h and 48 h.
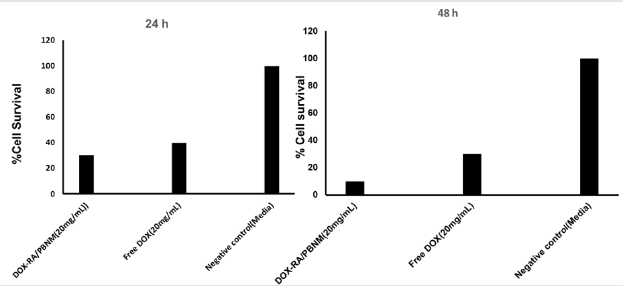
Cytotoxicity of DOX-RA-PBNM was evaluated in PC-3 cells using a CyquantTM cell proliferation assay kit, which emits fluorescence due to interactions between DNA and a special dye in the assay. Percentage of cell growth inhibition was measured by fluorescence caused by DNA of surviving cells after 48 h incubation. Doxorubicin concentration in the DOX-RA-PBNM formulation was adjusted to the same as that of free drug. DOX-RA-PBNM formulation showed significant cytotoxic effect in PC-3 cells after 48 h incubation (p < 0.05). It is worth noting that at 48 hours, the drug is not completely released from the nanomicelles (only 25%-40% released) as earlier depicted by the release profile. DOX-RA-PBNM nanomicelle formulation showed a significant difference in reduction of cell proliferation in PC-3 cells (p <0.05). Large amount of DOX from the DOX-RA-PBNM was delivered into PC-3 cells by endocytosis, causing high cytotoxic effect [38]. Direct impact of efflux mechanism on doxorubicin inside the cells may have been reduced by loading drug into the core of the DOX-RA-PBNM. Furthermore, lodging of DOX-RA-PBNM into the core of pentablock nanomicelles strongly enhanced the cytotoxic effect probably due to more drug molecules being ferried into the cells. Sustained drug release within the cell and reduction of efflux may have played a great role on effectiveness of the delivery system. The survival of cells l after incubation with DOX-RA/PBNM for 24 h and 48 h. The vulnerability of cancer cells to the treatment regime is related to the doxorubicin concentration.
After 48 h, it was observed the cell survival decreased with the increase in drug concentration. The cytotoxicity of DOX was almost the same as that of DOX-RA/PBNM at 24 h time point. However, depending on the increase in concentration of DOX-RA/PBNM, cytotoxicity continuously increased to a level much greater than that of pure doxorubicin, perhaps as a result of the delayed release of doxorubicin from pentablock nanomicelles (Figure 9). In general terms, long-term treatment with a drug where concentration gradually increases is much safer than an abrupt supply of high drug level [39]. This study demonstrates that delivery systems such as DOX-RA/PBNM may lower cytotoxicity or side effects caused by chemotherapy involving doxorubicin [6,39].
Conclusion
Stable DOX-RA/PBNM with mean diameters of 30nm ±5.0 nm was obtained. The release of doxorubicin from nanomicelles at pH 7.4 occurred slower, with approximately 40% of the total drug content being released within 108 h. This proved an adequate stability of doxorubicin in DOX-RA/PBNM at this pH, making it suitable for long circulation in an intravenous administration. DOX released much faster in acidic environments (pH 4.0) similar to that of tumors, allowing tumor specific drug release. FTIR and mass spectrometry analysis indicated that DOX and RA were present in the DOX-RA complex, which further confirmed the successful complex formation. The complex was perfectly lodged inside the nanomicelle core, which further reiterates that ion pairing of DOX with RA significantly raised the hydrophobicity of DOX. In vitro cell proliferation assay study showed that DOX-RA/PBNM had more toxicity compared with pure doxorubicin after 24 h as more drug molecules managed to enter the cells via the nanomicelles. The results of the release profile indicated that DOX-RA/PBNM had a sustained release from the nanomicelles hence the retention of high concentration of DOX which maximized the anticancer effect. The cytoplasmic distribution clearly indicates that DOX-RA/PBNM was taken up all over the cytoplasm and that the uptaken amount of DOX-RA/PBNM increased with time(time-dependent). This technique can reduce the systemic cytotoxicity and side effects caused by DOX therapy.
Acknowledgement
The authors would like to express their most sincere gratitude to 1) CAPES Foundation, Ministry of Education of Brazil, Brasília - DF 70040-020, Brazil for funding through SwB scholarship, 2) Ms. Barbara Fegley University of Kansas Medical Center, and 3) Mrs. Abrar Analfisah for FTIR facility.
References
- Patel KJ, Tredan O, Tannock IF (2013) Distribution of the anticancer drugs doxorubicin, mitoxantrone and topotecan in tumors and normal tissues. Cancer Chemother Pharmacol 72(1): 127-138.
- Luo JW, Zhang ZR, Gong T, Fu Y (2016) One-step self-assembled nanomicelles for improving the oral bioavailability of nimodipine. Int J Nanomedicine 11: 1051-1065.
- Primeau AJ, Rendon A, Hedley D, Lilge L, Tannock IF (2005) The distribution of the anticancer drug Doxorubicin in relation to blood vessels in solid tumors. Clinical cancer research: an official journal of the American Association for Cancer Research 11(24 Pt 1): 8782-8728.
- Zhao S, Minh LV, Li N, Garamus VM, Handge UA (2016) Doxorubicin hydrochloride-oleic acid conjugate loaded nanostructured lipid carriers for tumor specific drug release. Colloids Surf B Biointerfaces 145: 95-103.
- Wei WH, Dong XM, Liu CG (2015) In vitro investigation of self-assembled nanoparticles based on hyaluronic acid-deoxycholic acid conjugates for controlled release doxorubicin: effect of degree of substitution of deoxycholic acid. Int J Mol Sci 16(4): 7195-7209.
- Luk BT, Fang RH, Zhang L (2012) Lipid- and polymer-based nanostructures for cancer theranostics. Theranostics 2(12): 1117-1126.
- Andresen TL, Jensen SS, Jorgensen K (2005) Advanced strategies in liposomal cancer therapy: problems and prospects of active and tumor specific drug release. Prog Lipid Res 44(1): 68-97.
- Yatvin MB, Kreutz W, Horwitz BA, Shinitzky M (1980) pH-sensitive liposomes: possible clinical implications. Science 210(4475): 1253-1255.
- Mehnert W, Mader K (2001) Solid lipid nanoparticles: production, characterization and applications. Advanced drug delivery reviews 47(2-3): 165-196.
- Maia CS, Mehnert W, Schafer-Korting M (2000) Solid lipid nanoparticles as drug carriers for topical glucocorticoids. International journal of pharmaceutics 196(2): 165-167.
- Zur Muhlen A, Schwarz C, Mehnert W (1998) Solid lipid nanoparticles (SLN) for controlled drug delivery--drug release and release mechanism. Eur J Pharm Biopharm 45(2): 149-155.
- Schwarz C, Mehnert W (1999) Solid lipid nanoparticles (SLN) for controlled drug delivery. II. Drug incorporation and physicochemical characterization. Journal of microencapsulation 16(2): 205-213.
- Jores K, Haberland A, Wartewig S, Mader K, Mehnert W (2005) Solid lipid nanoparticles (SLN) and oil-loaded SLN studied by spectrofluorometry and Raman spectroscopy. Pharmaceutical research 22(11): 1887-1897.
- Wong HL, Bendayan R, Rauth AM, Wu XY (2004) Development of solid lipid nanoparticles containing ionically complexed chemotherapeutic drugs and chemosensitizers. J Pharm Sci 93(8): 1993-2008.
- Ma P, Dong X, Swadley CL, Gupte A, Leggas M (2009) Development of idarubicin and doxorubicin solid lipid nanoparticles to overcome Pgp-mediated multiple drug resistance in leukemia. J Biomed Nanotechnol 5(2): 151-161.
- Zhang J, Chen XG, Li YY, Liu CS (2007) Self-assembled nanoparticles based on hydrophobically modified chitosan as carriers for doxorubicin. Nanomedicine 3(4): 258-265.
- Vadlapudi AD, Cholkar K, Vadlapatla RK, Mitra AK (2014) Aqueous nanomicellar formulation for topical delivery of biotinylated lipid prodrug of acyclovir: formulation development and ocular biocompatibility. Journal of ocular pharmacology and therapeutics: the official journal of the Association for Ocular Pharmacology and Therapeutics 30(1): 49-58.
- Wei Z, Hao J, Yuan S, Li Y, Juan W, Sha X (2009) Paclitaxel-loaded Pluronic P123/F127 mixed polymeric micelles: formulation, optimization and In vitro characterization. International journal of pharmaceutics 376(1-2): 176-185.
- Agrahari V, Li G, Agrahari V, Navarro I, Perkumas K (2017) Pentablock copolymer dexamethasone nanoformulations elevate MYOC: In vitro liberation, activity and safety in human trabecular meshwork cells. Nanomedicine 12(16): 1911-1926.
- Agrahari V, Agrahari V, Mitra AK (2016) Nanocarrier fabrication and macromolecule drug delivery: challenges and opportunities. Therapeutic delivery 7(4): 257-278.
- Youm I, Agrahari V, Murowchick JB, Youan BB (2014) Uptake and cytotoxicity of docetaxel-loaded hyaluronic acid-grafted oily core nanocapsules in MDA-MB 231 cancer cells. Pharmaceutical research 31(9): 2439-2452.
- Xu J, Zhao Q, Jin Y, Qiu L (2014) High loading of hydrophilic/hydrophobic doxorubicin into polyphosphazene polymersome for breast cancer therapy. Nanomedicine 10(2): 349-358.
- Sawdon AJ, Peng CA (2014) Polymeric micelles for acyclovir drug delivery. Colloids Surf B Biointerfaces 122: 738-745.
- Lian H, He Z, Meng Z (2017) Rational design of hybrid nanomicelles integrating mucosal penetration and P-glycoprotein inhibition for efficient oral delivery of paclitaxel. Colloids Surf B Biointerfaces 155: 429-439.
- Christophersen OA (2012) Radiation protection following nuclear power accidents: a survey of putative mechanisms involved in the radioprotective actions of taurine during and after radiation exposure. Microb Ecol Health Dis, pp. 23.
- Youm I, Youan BB (2013) Uptake mechanism of furosemide-loaded pegylated nanoparticles by cochlear cell lines. Hear Res 304: 7-19.
- Jones LJ, Gray M, Yue ST, Haugland RP, Singer VL (2001) Sensitive determination of cell number using the CyQUANT (R) cell proliferation assay. J Immunol Methods 254(1-2): 85-98.
- Gou M, Gong C, Zhang J, Wang X, Wang X, Gu Y (2010) Polymeric matrix for drug delivery: honokiol-loaded PCL-PEG-PCL nanoparticles in PEG-PCL-PEG thermosensitive hydrogel. J Biomed Mater Res A 93(1): 219-226.
- Ding H, Wu F (2012) Image guided biodistribution of drugs and drug delivery. Theranostics 2(11): 1037-1039.
- Zhao Y, Duan S, Zeng X, Liu C, Davies NM (2012) Prodrug strategy for PSMA-targeted delivery of TGX-221 to prostate cancer cells. Molecular pharmaceutics 9(6): 1705-1716.
- Pearce AK, Simpson JD, Fletcher NL, Houston ZH, Fuchs AV (2017) Localised delivery of doxorubicin to prostate cancer cells through a PSMA-targeted hyperbranched polymer theranostic. Biomaterials 141: 330-339.
- Ben Jemaa A, Sallami S, Ceraline J, Oueslati R (2013) A novel regulation of PSMA and PSA expression by Q640X AR in 22Rv1 and LNCaP prostate cancer cells. Cell Biol Int 37(5): 464-470.
- Jin J, Sui B, Gou J, Liu J, Tang X, Xu H (2014) PSMA ligand conjugated PCL-PEG polymeric micelles targeted to prostate cancer cells. PloS one 9(11): e112200.
- Patil Y, Shmeeda H, Amitay Y, Ohana P, Kumar S (2018) Targeting of folate-conjugated liposomes with co-entrapped drugs to prostate cancer cells via prostate-specific membrane antigen (PSMA). Nanomedicine 14(4): 1407-1416.
- Wolf P, Gierschner D, Buhler P, Wetterauer U, Elsasser-Beile U (2006) A recombinant PSMA-specific single-chain immunotoxin has potent and selective toxicity against prostate cancer cells. Cancer Immunol Immunother 55(11): 1367-1373.
- Salgaller ML, Lodge PA, McLean JG, Tjoa BA (1998) Report of immune monitoring of prostate cancer patients undergoing T-cell therapy using dendritic cells pulsed with HLA-A2-specific peptides from prostate-specific membrane antigen (PSMA). Prostate 35(2): 144-151.
- Feldmann A, Arndt C, Bergmann R, Loff S, Cartellieri M (2017) Retargeting of T lymphocytes to PSCA- or PSMA positive prostate cancer cells using the novel modular chimeric antigen receptor platform technology "UniCAR". Oncotarget 8(19): 31368-31385.
- Yoo HS, Park TG (2001) Biodegradable polymeric micelles composed of doxorubicin conjugated PLGA-PEG block copolymer. J Control Release 70(1-2): 63-70.
- Bruge F, Damiani E, Puglia C, Offerta A, Armeni T (2013) Nanostructured lipid carriers loaded with CoQ10: effect on human dermal fibroblasts under normal and UVA-mediated oxidative conditions. International journal of pharmaceutics 455(1-2): 348-356.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...