
Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2644-1373
Research ArticleOpen Access
Comparative in Silico Analysis of Antigenic Proteins “Nucleocapsid Protein “of SARS-Cov-2 from Different Geographical Locations Volume 2 - Issue 4
Nada M Doleib1,2*
- 1Department of Biology, Faculty of Sciences and Arts, University of Jeddah, Saudi Arabia
- 2Department of Microbiology, Faculty of Applied and Industrial Science, University of Bahri, Sudan
Received: March 08, 2021 Published: March 15, 2021
Corresponding author: Nada M Doleib, Department of Biology, Faculty of Sciences and Arts,-Khulais, University of Jeddah, Saudi Arabia
DOI: 10.32474/LOJPCR.2021.02.000141
Abstract
SARS-CoV-2 Nucleocapsid protein considered as a vaccine target. Viral nucleocapsid protein is a potential antiviral drug target, serving multiple critical functions during the viral life cycle. However, the structural information of SARS-CoV-2 in different geographical locations considered as an important factor in order to produce a global vaccine for the pandemic. The present investigation relies on the analysis of the similarities and differences of the SARS-CoV-2 Nucleocapsid protein from various geographical locations, considering certain protein parameters, as well as the study of the alignment of the protein sequence of amino acids.
Keywords: Nucleocapsid protein; SARS-CoV-2; geographical locations; bioinformatics
Introduction
SARS-CoV-2 is a single-stranded, positive-sense RNA virus with a 30 kb genome, one of the largest among RNA viruses [1]. The viral envelop of SARS-CoV-2, contains many proteins such as spike protein, and glycoprotein [2]. Another protein incorporated with the viral genome is nucleocapsid protein which is responsible for protection the virus from the host cell environment [3]. The SARSCoV- 2 spike protein is being used as the principal antigen target in vaccine progress. However, the multifaceted molecular details of viral entrance may lead to obstacles with the vaccine reaction [4]. The nucleocapsid (N) protein is a significant antigen for coronavirus, which contribute to RNA package and virus particle discharge [5]. After infection, the N protein enters the host cell along with the viral RNA to facilitate its replication and to process the assembly and release of the virus. SARS-CoV N comprises two distinct RNA-binding domains (N-terminal domain and C-terminal domain) connected by a poorly structured linkage region. Due to positive amino acids, SARS-CoV N-terminal domain and C-terminal domain have been documented to bind to the viral RNA genome [6]. Serological diagnosis has shown that the unique antibodies to the N protein in the serum of SARS patients have a higher sensitivity and longer persistence than those of other structural SARS-CoV proteins. In addition, anti-N antibodies have been observed at an early stage of infection with a high specificity. Thus, any information obtained from the study of this protein, whether in vivo or in vitro, will improve our understanding of COVID-19 and enable us to develop better biological agents for the treatment or diagnosis of diseases [7]. It is becoming clearer how important this protein is, to the multiple stages of the viral life cycle. This studie provide valuable and timely insights specific to the SARS-CoV-2 N protein, a vaccine target that has some distinct advantages over other possible SARSCoV- 2 antigens according to geographical location. Because of the conservation of the N-protein sequence, the increased awareness of its genetics and biochemistry regarding different geographical location, the N-protein SARS-CoV-2 should be considered as a global vaccine candidate for SARS-CoV-2. The present investigation relies on study the similarities and difference of Nucleocapsid protein “of SARS-CoV-2 from different geographical locations considering some protein parameters as well as study the alignment of amino acid sequence of protein.
Materials and Methods
Sequences, alignment, and construction of phylogenetic tree
Amino acids sequences for the Nucleocapsid protein of SARSCoV- 2 from different geographical locations were obtained from the National Center for Biotechnology Information database. The accession numbers of the corresponding database entries and isolation source are listed in Table 1. Multiple sequence alignment of Nucleocapsid protein of SARS-CoV-2 was performed in order to find the evolutionary relationships between sequences from different geographical location to identify shared patterns. Sequences were multiply aligned Jalview software version 2.8 [8]. Phylogenetic trees were constructed with neighbour-joining using MEGA [9]. A distance matrix was generated using the model building Jones-Taylor-Thornton [10]. Graphical way of representing and visualizing consensus data developed of amino acid multiple sequence alignment developed were displayed according to method described by Tom Schneider and Mike Stephens [11].
Table 1: Accession numbers of NCBI entries for Nucleocapsid protein of SARS-CoV-2 from differen geographical locations.
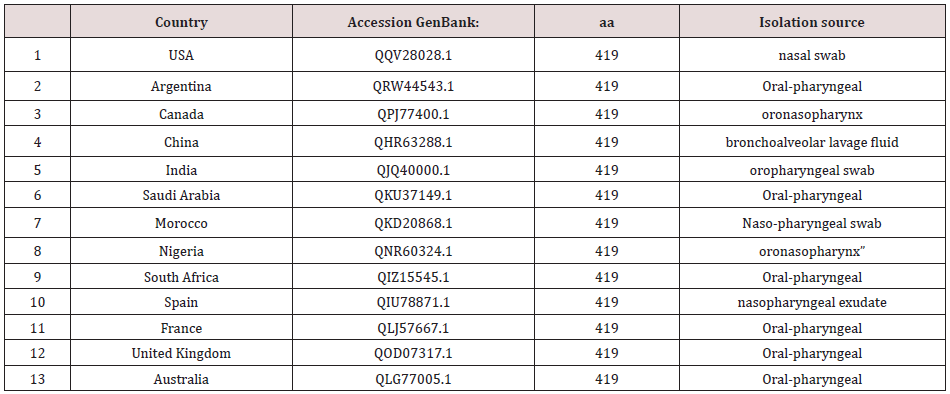
*aa is arrangement of amino acids in a protein.
Computation of amino acid composition and molecular weight of protein sequences
Estimation of the amino acid composition and molecular weight was determined using Isoelectric Point Calculator (IPC), a web service and a standalone program for the accurate estimation of protein and peptide characteristic [12].
Solvent content of protein crystals
From the currently available sequence, of the solvent content of Nucleocapsid protein. The fraction of the crystal volume occupied by solvent was calculated according to Matthews [13].
Results
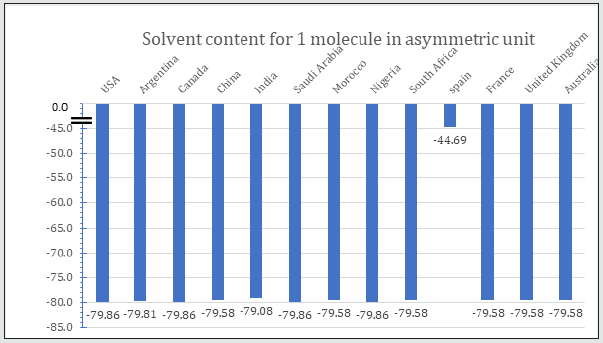
Table 2 is displayed the chemical composition of the Nucleocapsid protein “of SARS-CoV-2 from different geographic location. The preset data revealed that amino acid composition of the tested 13 sequences showed high similarity except for the protein sequence from Spain. Figure 1 represented the overall height of the stack indicates the amino acid sequence conservation at each position of protein, while the height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. In general, sequence logo of Nucleocapsid protein provides that almost the sequence from different location did not differ from each other and revealed high similarity. Regarding Phylogeny estimation for Nucleocapsid protein of SARS-CoV-2 from different geographical locations. Neighbour-joining tree constructed using Mega 10.1.8. The multiple sequence alignments are depicted in Figure 2. Constructed phylogenetic tree is depicted in Figure 3. The current results indicated that; Neighbour-joining tree of Nucleocapsid protein of SARS-CoV-2 displays the comparison of amino acid sequences of Nucleocapsid protein from different location. The results shows that the phylogenetic analysis rooted by two clusters (Figure 3). Cluster 1 represents France, Australia, China, India, South Africa, Morocco, United Kingdom and Spain. In the Cluster 1 Spain considered as the main root. Cluster 2 represents USA, Canada, Argentina, Saudi Arabia and Nigeria. The main root of the cluster 2 is Canada. Figure 4 shows Matthews coefficient , the current results revealed that the parameter value equal to 0.68 for all locations except for Spain its value reaches 0.85. Figure 5 shows the results for Solvent content of protein crystals were near 80 % for all geographical locations but has been observed that the value calculate for Spain was the lowest equal to 44%. As showed in Table 3 molecular weight of Nucleocapsid protein “of SARS-CoV-2 from different geographic location ranges from 36759.93 to 45696.7 Dalton, exhibited low variability in molecular weight.
Figure 1: Multiple sequence alignment of Nucleocapsid protein “of SARS-CoV-2 from different geographic location.
The amino acid sequences were grabbed from NCBI (National Center for Biotechnology Information) database.
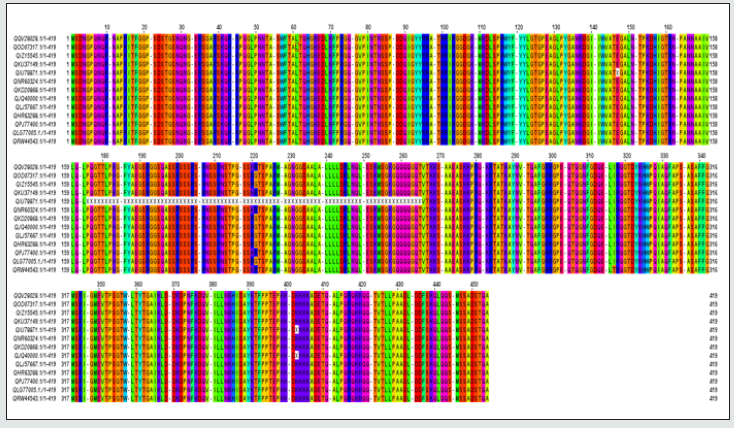
Table 2: Chemical composition of the Nucleocapsid protein “of SARS-CoV-2 from different geographic location.
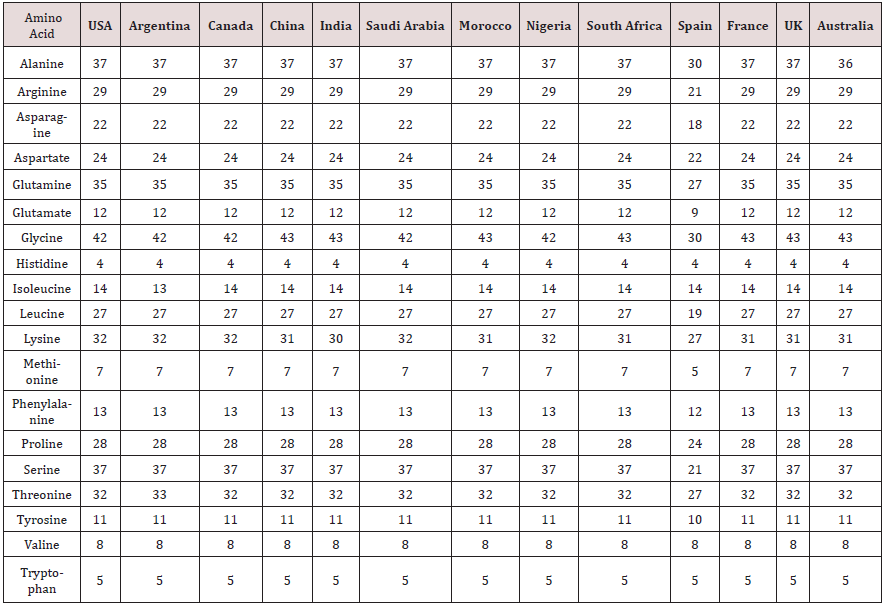
Figure 2: Representation of a scoring matrix constructed for a multiple sequence alignment of Nucleocapsid protein
“of SARS-CoV-2 from different geographic location. Each of the 20 amino acids frequently found in proteins is
assumed a mark for each location in the sequence agreeing to the frequency with which they occur in the original
alignment.

Figure 3: Phylogenetic analysis of protein sequences Nucleocapsid protein “of SARS-CoV-2 from different geographic
location. Bootstrap percentage values as obtained from 100 resampling of the date set are given at the nodes of the
tree. The obtained tree calculated by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances
estimated using the Jones-Taylor-Thornton model.
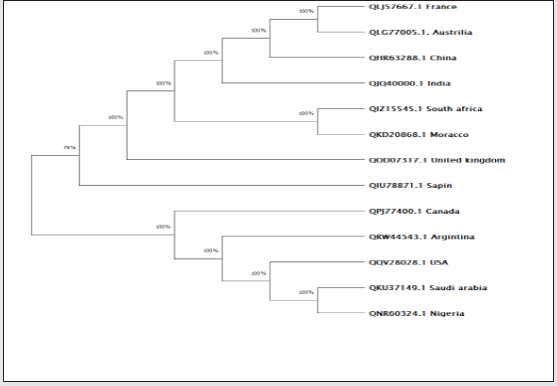
Table 3: Molecular weight of Nucleocapsid protein “of SARS-CoV-2 from different geographic location.
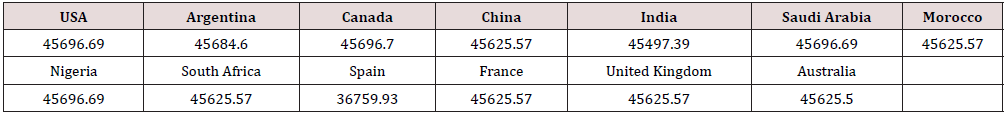
Figure 4: Matthew’s coefficient for Nucleocapsid protein of SARS-CoV-2 from different geographical locations.
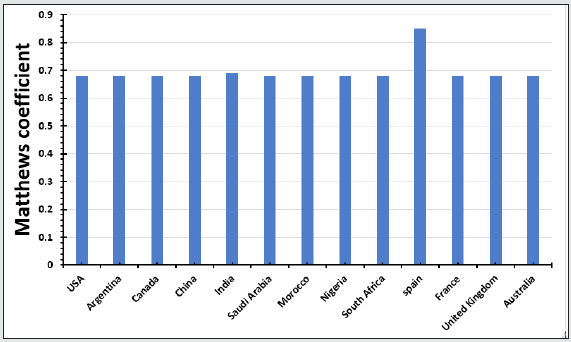
Figure 5: Solvent content for Nucleocapsid protein of SARS-CoV-2 from different geographical locations.br>
Discussion
The current study demonstrated that the Nucleocapsid protein
analysis from different location shared a common primary amino
acid composition which resulted in a uniform sequence alignment.
Previously reports declared that alignments are a powerful way to
compare related protein sequences. They can be used to capture
various facts about the sequences aligned, such as communal
evolutionary descent or shared structural function [14]. The obtain
composition information of SARS-CoV-2 Nucleocapsid protein
revealed high similarity in different geographic location with mild
variability in amino acid concentration. The study compared aminoacid
distributions between different locations. When looking at
overall amino acid frequencies it could be recognized that byand-
large, amino acid frequencies in all investigated sequences
mirrored to each other except for the sequence obtained from
Spain. The biggest differences arose in all amino acids except for
Histidine, Isoleucine, Tryptophan and valine. According to Jackson
et al. [15] there are several patterns of sequence variation that are
consistently seen in natural proteins.
The first steps in a macromolecular structure determination of
protein are to detect the count of molecules in the crystallographic
asymmetric unit. The crystal volume per unit of protein molecular
weight, known as Matthews coefficient. A substantial percentage
of the protein crystals volume is employed by solvent ranged from
27% to 78%, with the most common value being about 43% [16].
The Matthews Coefficient and solvent content are calculated for
Nucleocapsid protein “of SARS-CoV-2 from different geographic
location which revealed almost the same results or all locations
except for Spain. Water plays an important role in the structure
of biomolecules and often influences protein function [17]. Water
molecules not only affect protein folding, but also mediate biological
processes such as enzymatic reactions and molecular recognition.
Information about the fraction of water (solvent) plays a significant
role in the X-ray structure determination process [18]. The present results revealed that solvent content for Nucleocapsid protein of
SARS-CoV-2 from different geographical locations were almost
similar except for Spain.
Conclusion
In the current study, the variation of Nucleocapsid protein of
SARS-CoV-2 composition and its alignment were investigated for
different location, it revealed high similarity for each other and
showed a uniform sequence alignment with mild variability in
amino acid concentration. The crystal volume per unit of protein
molecular weight of Nucleocapsid protein “of SARS-CoV-2 revealed
almost the same results or all locations except for Spain.
Conflicts of Interest: The authors declare no conflict of interest.
References
- Savastano A, Ibáñez de Opakua A, Rankovic M, Zweckstetter M (2020) Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun 11(1): 1-10.
- Henderson R, Edwards RJ, Mansouri K, Janowska K, Stalls V, et al. (2020) Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat Struct Mol 27(10): 925-933.
- Zivcec M, Safronetz D, Scott DP, Robertson S, Feldmann H (2018) Nucleocapsid protein-based vaccine provides protection in mice against lethal Crimean-Congo hemorrhagic fever virus challenge. Bente DA, editor. PLoS Negl Trop Dis 12(7): e0006628.
- Dutta NK, Mazumdar K, Gordy JT (2020) The Nucleocapsid Protein of SARS-CoV-2: a Target for Vaccine Development. J Virol 94(13): 647-720.
- Zeng W, Liu G, Ma H, Zhao D, Yang Y, et al. (2020) Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun 527(3): 618-623.
- Fan H, Ooi A, Tan YW, Wang S, Fang S, et al. (2005) The nucleocapsid protein of coronavirus infectious bronchitis virus: Crystal structure of its N-terminal domain and multimerization properties. Structure 13(12): 1859-1868.
- Sethuraman N, Jeremiah SS, Ryo A (2020) Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 323(22): 2249-2251.
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9): 1189-1191.
- Hall BG (2013) Building Phylogenetic Trees from Molecular Data with MEGA. Mol Biol Evol 30(5): 1229-1235.
- Munjal G, Hanmandlu M, Srivastava S (2020) Phylogenetics Algorithms and Applications. In: Advances in Intelligent Systems and Computing. Springer Verlagp pp. 187-94.
- (2005) WebLogo - About.
- Kozlowski LP (2016) IPC - Isoelectric Point Calculator. Biol Direct 11(1): 50-55.
- Matthews BW (1968) Solvent content of protein crystals. Journal of Molecular Biology 33(2): 491-497.
- Reilly C (2020) Sequence Alignment. In: Statistics in Human Genetics and Molecular Biology. pp. 125-138.
- Jackson EL, Ollikainen N, Covert AW, Kortemme T, Wilke CO (2013) Amino-acid site variability among natural and designed proteins. PeerJ 1: 211.
- Kantardjieff KA, Rupp B (2003) Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci 12(9): 1865-1871.
- Privalov PL, Crane-Robinson C (2017) Role of water in the formation of macromolecular structures. Eur Biophys J 46(3): 203-224.
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, et al. (200) Folding, Modification, and Degradation of Proteins.




