
Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2644-1373
Research ArticleOpen Access
A De-Novo SCN2A Mutation Identified in a Chinese Patient With Epilepsy: A Case Report Volume 2 - Issue 3
>Shuna Duan1**, Weicheng Hao1**, Yongxiong Zhao1, Xiuping Zhang1, Guihua An1, Jianbing Zhang1, Congxi Liu1, Yaxi Zhang2, Yanxia Mi2, Jianli Zhang2, Yuxi Liu1* and Jianxiong Duan2*
- 1Department of Neurology, the Second Hospital of Shanxi Medical University, Taiyuan City, People’s Republic of China
- 2Shanxi Guoxin Caregeno Medical Laboratory, People’s Republic of China
- ** Co First author: These authors contributed equally to this work.
Received: January 15, 2021 Published: January 25, 2021
Corresponding author: Jianxiong Duan (jianxiongDuan@careegeno.com) Yuxi Liu (liuyuxi2003@163.com)
DOI: 10.32474/LOJPCR.2021.02.000139
Abstract
Objective: To investigate the genetic causes of epilepsy in a 5-year-old Chinese female patient.
Methods: Clinical diagnosis and next-generation sequencing.
Results: The patient carries a de-novo heterozygous missense mutation (c.3686 A>T p.Asp1229Val) in the SCN2A gene. This mutation was evaluated as a pathogenic mutation based on the standards and guidelines of ACMG (American College of Medical Genetics and Genomics) and clinical research publications.
Conclusion: The de-novo heterozygous mutation (c.3686 A>T p.Asp1229Val) in the SCN2A gene is the genetic cause of the epilepsy for the patient. So far, this mutation of SCN2A gene is the first reported in the worldwide overall populations.
Keywords: Chinese; Epilepsy; SCN2A gene
Abbreviations: Whole Exome Sequencing (WES); Sanger sequencing; American College of Medical Genetics and Genomics (ACMG)
Introduction
The SCN2A gene encodes the voltage-gated sodium channel Na(v)1.2, which plays an important role in the initiation and conduction of action potentials. SCN2A is expressed in axon initiation segments and at nodes of Ranvier in myelinated nerve fibers during early development, and is later expressed in unmyelinated axons [1]. Mutations in the SCN2A gene, cause a variety of neuropsychiatric syndromes with different severity ranging from self-limiting epilepsies with early onset to developmental and epileptic encephalopathy with early or late onset and intellectual disability (ID), as well as ID or autism without seizures [2-4]. So far, more than 700 mutations in SCN2A were reported to be associated with epilepy [5]. The aim of the present study was to detect and report genetic causes of a 5-year-old Chinese female patient with epilepsy. The patient was found to have a de-nove mutation (c.3686 A>T p.Asp1229Val) in the SCN2A gene that has not reported in the previous studies.
Materials and Methods
Clinical diagnosis
A 5-year-old female Chinese patient who was born with no family history of seizures or other types of neurological diseases, has experienced epileptic spasms since she was 1.5 years old. The patient had fallen down while she was walking, and her head contacted the ground, but she did not loss consciousness at the time. On the same day, the patient began to nod her head continuously, with each occurance lasting approximately several seconds in length. Since then, her head nods occurred in clusters daily, and up to 30 to 40 times a day at the worst cases. The patient was carried to the hospital and was diagnosed with infantile spasms. Her symptoms of head nodding was relieved after treatment with intravenous immunoglobulin (pH4), Sodium valproate oral solution and Topiramate. The patient took Sodium valproate oral solution and Topiramate after her discharge from the hospital. However, she still had the head nods which occurred at random intervals. After she turned 2-years-old, the head nods did not occur again, but she began to have epilepsy symptoms such as loss of consciousness, twitching of the limbs, spitting out white foam et al. The epilepsy happened approximately once a month and lasted approximately 1 to 2 minutes in length at each occurence. The patient visited the hospital again and the Clonazepam was added to her medications [6]. Her symptoms of epilepsy were being controlled well and did not happen within a 8 month period. However, in April of 2019, when the patient was 5 years old, her head nods occurred again without any obvious causes. This time, the head nods happened in the early morning, and 4~5 times everyday. The patient was admitted to our hospital (the Second Hospital of Shanxi Medical University). The following tests were performed for the patient: physical examinations, brain MRI, and electroencephalogram.
Molecular test
In order to study the cause of the disease, whole exome sequencing was performed for the patient. Furthermore, Sanger sequencing was used to verify the variant for the patient and her parents. Sequencing data was analyzed by using numerous bioinformatics’ softwares, the pathogenicity of the mutation was evaluated based on the standards and guidelines of ACMG (American College of Medical Genetics and Genomics), Clinvar database, OMIM (Online Mendelian Inheritance in Man), HGMD (Human Gene Mutation Database), and clinical research papers published in scientific journals.
Results and Analysis
Clinical data analysis
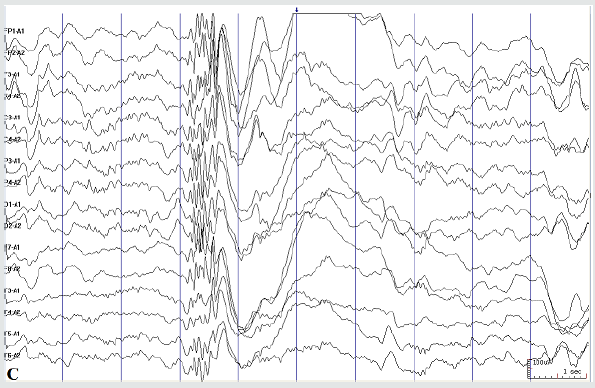
The patient was admitted to our hospital in April of 2019. The patient appeared normal under physical examinations. She has a clear mind, but only can pronunce simple words. She had poor orientation, cognition, memory, calculation, and attention span. Brain MRI showed that the symmetrical brain hemispheres on both sides, and the structures of gray and white matter were normal. No other abnormalities were found in the brain parenchyma (Figure 1). The electroencephalogram of the patient showed that the slowwave activity were in the awake period (Figure 2A). Total epileptic discharge during sleep period (Figures 2B & 2C). The patient was given intravenous human immunoglobulin and Dexamethasone for immunotherapy. The medications she has had were also adjusted to Levetiracetam tablets, Clonazepam, Topiramate, and Sodium valproate oral liquid. No recurrence of the head nods after 3 month of the treatment. Figure 2D showed that no obvious abnormal discharge was found in the patient’s return visit on Oct. 2020.
Figure 1: Head MRI (right and left): symmetrical brain hemispheres on both sides, and the structures of gray and white matter were normal.
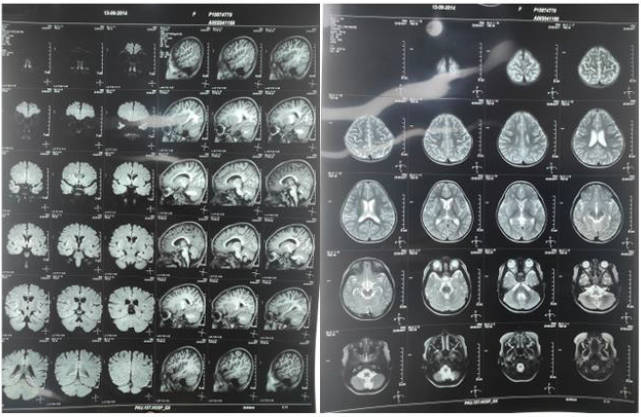
Figure 2A: Seizure-free interval: Slow-wave activity during the awake period (A hospital visit on Apr. 2019).

Figure 2B: Seizure-free interval: spike slow complex waves and polyspike slow complex waves during sleep period (A
hospital visit on Apr. 2019).
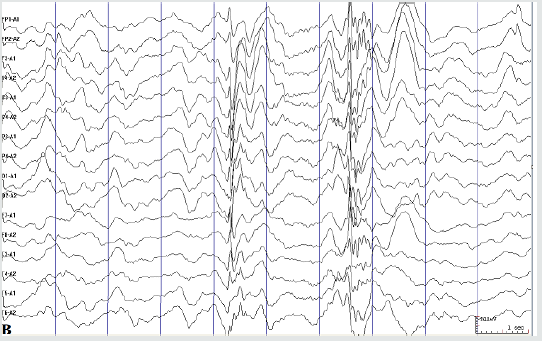
Figure 2C: Seizure-free interval: spike wave burst during sleep period (A hospital visit on Apr. 2019)
Molecular biological data analysis
In order to identify the causes of the patient’s seizures, we conducted Whole Exome Sequencing (WES) based on Nextgeneration sequencing for the patient. a heterozygous missense mutation in SCN2A gene (c.3686 A>T p.Asp1229Val) reference transcript, NM_001040143) was detected in the patient. This variant was further confirmed by Sanger Sequencing, and not detected in either of the healthy parents, which indicated that this variant was de novo (Figure 3A). No other pathogenic variants were detected in more than approximately 800 genes that were defined by the OMIM database as related with epilepsy syndromes for this patient (Figures 3B & 3C). The mutation c.3686 A>T (p.Asp1229Val) either not being recorded in any clincial disease-related database (Clinvar and HGMD), nor in Human genome databases (1000 Genome and Genome mutation frequency database). The function prediction databases (SIFT and polyphen, etc.) predicted this mutation to be damaging. This variant was evaluated as a pathogenic mutation based on the standards and guidelines of ACMG.
Figure 3A: Patient’s result (AT) : Sanger sequencing of the SCN2A gene mutation (c.3686 A>T p.Asp1229Val). sequencing
result of the patient showed a heterozygous A-to-T de novo transition (red arrow) at codon 3686 resulting in amino acid
exchange (A->T; Asp -> Val).
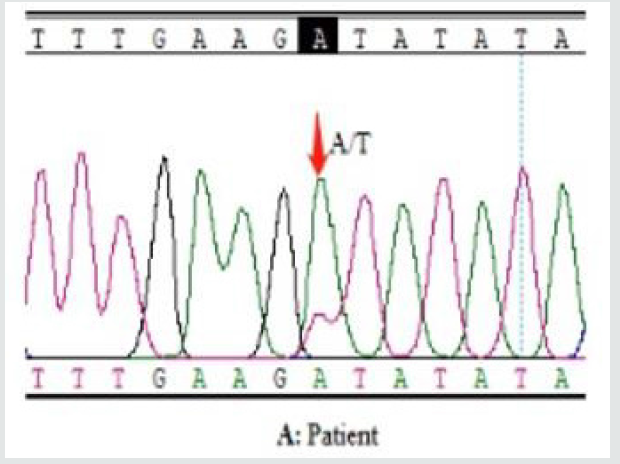
Figure 3B: Sanger sequencing of the SCN2A gene mutation (c.3686 A>T p.Asp1229Val). Wild-type sequence in patient’s
mother and father (red arrow). Patient’s mother (AA).
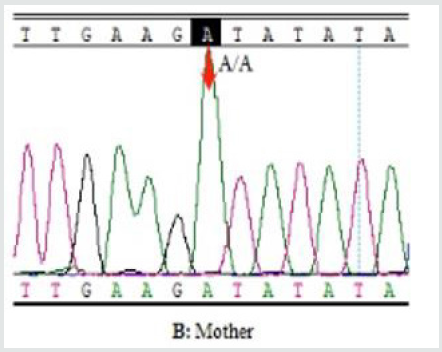
Figure 3C: Sanger sequencing of the SCN2A gene mutation (c.3686 A>T p.Asp1229Val). Wild-type sequence in patient’s
mother and father (red arrow). Patient’s father (AA).

Discussion
Pathogenic variants in SCN2A are reported in a spectrum
of neurodevelopmental disorders including developmental and
epileptic encephalopathies, benign familial neonatal-infantile
seizures, episodic ataxia, and autism spectrum disorder and
intellectual disability with and without seizures [1]. More than
1000 mutaions of SCN1A gene were reported in the Clinvar
database, including deletion, duplication, indel, insertion and single
nucleotide types. Around 70% of those variations was evaluated as
responsible for the occurences of Benign familial infantile seizures
or early infantile epileptic encephalopathy 11. The single nucleotide
mutations were the most frequently reported mutations and were
84% in the total mutation types. However, only about 8.5% of the
single nucleotide mutations were due to de-nove mutations in the
SCN2A gene. Comparing with the 65% of the de-novo rate in the
SCN1A gene, the rate of de-novo in the SCN2A gene is significantly
lower [6]. We evaluated WES data from 332 Chinese patients with
epilepsy and identified the one de-nove missense mutation in the
SCN2A in this patient and the mutation was evaluated as the cause
of the disease. Generally, the de-novo mutations of SCN2A gene
often lead to severe phenotype with developmental delay [3]. The
patient in our case reported here is only 5 years old, but she had
epilepsy syndromes for more than three years and had more severe
symptoms such as poor orientation, cognition, memory, calculation,
and attention et al.
The mutation of the SCN2A gene we detected (c.3686 A>T
p.Asp1229Val) has not been recorded in the Clinvar database as
of today [6]. Our report provided further evidence for the cause
of epilepsy from a genetic level. We predict the result will be
immediately useful for the clinical interpretation of SCN2A variants,
and also provides deeper insights for SCN2A mutations associated
with the broad clinical spectrum of seizures.
Declarations
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of the Second Hospital of Shanxi Medical University. Written informed consent was obtained from individual or guardian participants. This study was supported in the part by grants from the Epilepsy Research Fund (UCB No. 2014007) from China antiepileptic Association.
References
- Reynolds C, King MD, Gorman KM (2020) The phenotypic spectrum of SCN2A-related epilepsy. Eur J Paediatr Neurol 24:117-122.
- Hedrich UBS, Lauxmann S, Lerche H (2019) SCN2A channelopathies: Mechanisms and models. Epilepsia 60 Suppl 3:S68-S76.
- Q Zeng, YH Zhang, XL Yang, J Zhang, AJ Liu, et al. (2018) Phenotype study of SCN2A gene related epilepsy. Zhonghua Er Ke Za Zhi 56(7):518-523.
- Stephan J Sanders, Arthur J Campbell, Jeffrey R Cottrell, Rikke S Moller, Florence F Wagner, et al. (2019) Progress in understanding and treating SCN2A-mediated disorders. Trends Neurosci 41(7):442-456.
- Wolff M, Johannesen KM, Hedrich UBS, Masnada S, Rubboli G, et al. (2017) Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain 140(5):1316-1336.
- Clinvar.





