Lupine Publishers Group
Lupine Publishers
Menu
Research Article(ISSN: 2770-5447) 
Identifying Long Non-Coding RNAs Associated with Acute Myocardial Infarction Volume 2 - Issue 3
Fanyan Luo1*, Lizhi Lv1, Weijie Ye2 and Rong Liu2
- 1Department of Cardiothoracic Surgery, Central South University, PR China
- 2Department of Clinical Pharmacology, Central South University, PR China
Received: October 01, 2019; Published: October 11, 2019
Corresponding author: Fanyan Luo, Department of Cardiothoracic Surgery, Xiangya Hospital, Central South University, Changsha 410008, PR China
DOI: 10.32474/ACR.2019.02.000139
Abstract
Background: Accumulated evidence suggests that dysregulated expression of long non-coding RNAs (lnc RNAs) may participate in the development of cardiovascular diseases. In this study, we aim at identifying circulating lnc RNAs associated with acute myocardial infarction (AMI).
Materials and methods: By repurposing microarray probes from two public datasets (GSE48080, GSE66360) from gene expression omnibus database, an array-based transcriptional analysis of lnc RNAs in AMI patients and controls were conducted by us. Data analyses with R and Bioconductor.
Results: Six lnc RNAs (MIR22HG, RP11-296O14.3, IDI2-AS1, RP11-539L10.2, MIR3945HG, RP11-96D1.11) were identified to be expression differently in AMI (Bonferroni p value <0.01), and a distinguish score was constructed based on the expression data of two lnc RNAs (RP11-539L10.2 and MIR22HG). This distinguish score showed predictive power in distinguishing AMI from controls in the training (AUC=0.92) and validating (AUC=0.70) datasets. Functional enrichment analyses revealed potential functional roles of MIR3945HG in immune response.
Conclusion: Taken together, our newly identified circulating lnc RNAs may have a potential role in the development of AMI.
Keywords: long non-coding RNA, myocardial infarction; data mining, biomarker, prediction model.
List of Abbreviations: Lnc RNAs: Long Non-coding RNAs; AMI: Acute Myocardial Infarction; CVD: Cardiovascular Disease; MI: Myocardial Infarction; GEO: Gene Expression Omnibus; AUC: Area Under The Curve; PCGs: Protein Coding Genes; ROC: Receiver Operating Characteristic Curve; GO: Gene Ontology
Background
Cardiovascular disease (CVD) is the leading cause of death worldwide, with the 2013 global study illuminate that CVD is responsible for 17.3 million deaths globally [1]. It brings about 31.5% of all deaths, 45% of all non-communicable disease deaths, and twice more than that caused by cancer [2]. In most regions of the world, the age-standardized of myocardial infarction (MI) has decreased over past two decades, and the global morbidity of MI has increased by 29 million disability-adjusted life years [3]. Despite the significant advancement of pharmacotherapy, revascularization strategies and organ transplantation, the main cause of death in adult above 35 years old is MI in the United States [4]. Some assessments of cardiovascular risk factors such as hypertension, diabetes, and smoking play vital roles for doctor to prevent and predict disease [5-7]. Further, advances in genomics and proteomics have promoted the development of novel molecular biomarkers which have potential clinical values for AMI [1-8]. In recent studies, long non-coding RNAs (lnc RNAs) have attracted great interest in the domain of cardiovascular diseases [9]. Lnc RNAs, range from 200 nucleotides (bp) to multiple kilobases (kb), are mRNA-like transcripts but lack of protein coding capacity [10]. Linc RNAs play a vital role in some biological processes, such as epigenetic and post-transcription regulation. Dysregulated lnc RNAs participate in regulating cardiac development and in the pathogenesis of heart failure [11,12].
For example, lncRNA ANRIL (also known as CDKN2BAS) is associated with the risk of coronary atherosclerosis [13], peripheral artery diseas [14], carotid arteriosclerosis [15], and other vascular disease. It has been reported that expression levels of lnc RNAs are altered in the cardiac tissue [16] and blood [17] after AMI. Zhang et al measured the circulating levels of 15 cardiovascular disease related lnc RNAs, and found that circulating lncRNA ZFAS1 and CDR1 are predictive of AMI [18]. By data mining previously published gene expression microarray data from database such as gene expression omnibus (GEO) and Array Express, we can get the lncRNA profiling since thousands of lncRNA-specific probes were represented on the commonly used microarray platforms such as Affymetrix Human U133 Plus 2.0 arrays [19]. In this study, we applied this method to conduct gene expressions of lnc RNAs profiling on two cohorts from GEO database. We investigated the expression of lnc RNAs in AMI patients and control subjects. A three-lncRNA signature were identified from the GSE66360 test series showed predictive power in distinguishing AMI from controls and validated it in the GSE48060 validation series. We also integrated with proteincoding mRNA expression data to predict the potential role of our identified lnc RNAs.
Materials and Methods
Datasets utilized in our study
Two datasets with the profiling data of gene expression were downloaded from the GEO database (http://www.ncbi.nlm.nih. gov/geo/) with accession number GSE66360 and GSE48060 [1], respectively. In the training dataset GSE66360, circulating endothelial cells from patients experienced acute myocardial infarction (AMI, n=49) and healthy controls (n=50) were isolated and gene expression pattern was determined by using the Affymetrix Human U133 Plus 2.0 arrays (Affymetrix). In the validating dataset GSE48060, the study samples consisted of whole blood from 31 first-time AMI patients and 21 controls with a normal echocardiogram. Nucleated cells, fractionated from heparinized blood, were isolated and gene expression pattern was determined by using the Affymetrix Gene Chip Human Genome U133 Plus 2.0, which includes 54,675 probe sets.
Microarray processing and lncRNA expression mining
The probe sets of Affymetrix Human U133 Plus 2.0 arrays that were not mapped for pseudogene transcripts or protein-coding transcripts but were uniquely and perfectly assigned for lncRNA sequences were obtained from Du’s study [20] (http://cistrome. org/lncRNA/lncRNA_data_repository.html, file Array. probe. alignment/U133p2.lncRNA.uniq). Every lncRNA was confirmed by at least four probes. To reduce the probability of inaccurate annotations, the lncRNAs obtained from our analysis and lncRNAs defined in the GENCODE project (http://www.gencodegenes.org/, release 25) [21] were cross-referenced by Ensembl id. Finally, up to 2653 probes corresponding to 2183 lncRNAs were left. The CEL files were normalized with the MAS5 algorithm using the “affy” R Bioconductor package (http://www.bioconductor.org/packages/ release/bioc/html/affy.html). Furthermore, the probe-level expression profiles were converted into lncRNA-based expressions with the collapse row function [22], specifically, when multiple probes assigned in one lncRNA, the expression level of the lncRNA was calculated with the mean expression level of those probes. Finally, the lncRNA expression levels were normalized with a mean of 0 and an SD of 1.
Statistical Analysis
To identify AMI associated lnc RNAs, we conducted t test to assess the relationship between the continuous expression level of each lncRNA in AMI and health controls. The lnc RNAs with Bonferroni p values less than 0.01 were considered to be statistically significant and associated with AMI. Multivariate logistic regression was performed for the above selected lnc RNAs, and those lnc RNAs with a multivariate model p value of less than 0.05 were left for the distinguish score calculation. The distinguish score was calculated to evaluate each patient’s probability of AMI according to the following formula:
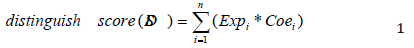
where n is the number of lnc RNAs in the model; Expi stands for the expression level of lnc RNAi; Coei represents the estimated regression coefficient of lnc RNAi in the multivariable logistic regression model. Patients who have higher distinguish scores are expected to have a higher probability of AMI. The area under the receiver operator characteristic curve (AUC) was used to access the classification performance of the distinguish scores according to their capability to distinguish AMI from normal control. Moreover, UC value was calculated via ROCR R package (https://cran.r-project. org/web/packages/ROCR/index.html). All statistical analyses in this study were performed using the R statistical software version 3.3.3 [23] and Bioconductor with related packages.
Functional Prediction of Lnc RNAs
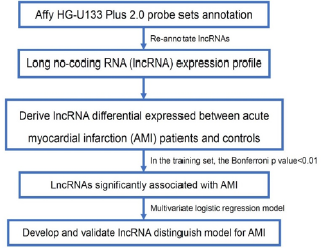
A previous study reported that the biological functions of lnc RNAs are correlated with the co-expressed protein coding genes (PCGs) [24]. Thus, we tested the correlation between the expression levels of each paired lncRNA and PCG. The PCGs was defined as lncRNA correlated when the correlation coefficient was higher than 0.4 in both the datasets. The GO biological process (GOTERMBP- ALL) enrichment analyses of the PCGs co-expressed with AMI associated lnc RNAs were performed to predict the function of AMI associated lnc RNAs via the DAVID annotation tool (http://david. abcc.ncifcrf.gov/) with the functional annotation clustering option [25]. The enriched Gene Ontology (GO) terms with a Bonferroni p value of <0.05 were considered as a potential function of AMI associated lnc RNAs. The significantly enriched GO terms were visualized with the Enrichment Map Plugin in Cityscape [26]. The overall workflow of this study is shown in Figure 1.
Results
Characteristic of the patients
Two datasets were downloaded from GEO with the following accession numbers: GSE66360 and GSE48060. A total of 151 samples with 99 individuals for the GSE66360 (49 AMI cases, 50 controls) and 52 individuals for the GSE48060 (31 AMI cases, 21 controls) were analyzed (Table 1).
Table 1: Patient characteristics of the datasets utilized in our study.
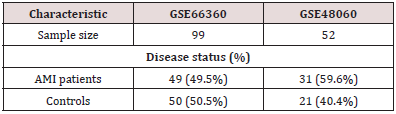
AMI: acute myocardial infarction
Identification of lnc RNAs from GSE66360
The GSE6630 (n=99) was selected as training dataset to determine the association between lnc RNAs and AMI. Six lnc RNAs (MIR22HG, RP11-296O14.3, IDI2-AS1, RP11-539L10.2, MIR3945HG, RP11-96D1.11) were found to be significantly associated with AMI patient (Bonferroni <0.01, Table 2) using differential expression analysis. Among these above six lnc RNAs, the expression of RP11-296O14.3, RP11-539L10.2 and RP11- 96D1.11 were significant lower in AMI patients that controls. Meanwhile, the expression of the remaining three lnc RNAs (MIR22HG, MIR3945HG, and IDI2-AS1) were significant higher in AMI than controls. Lnc RNAs with p value than 0.05 were listed in Table 1.
Table 2: Logistic regression model for myocardial infarction in patients with complete clinical and genomic data in the training dataset (n=99).
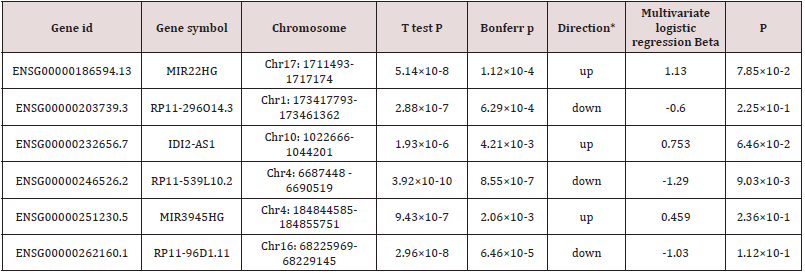
*The value of direction up and down suggest the expression of this gene significant higher and lower in AMI patients that controls, respectively.
Two-lncRNA signature and myocardial infarction
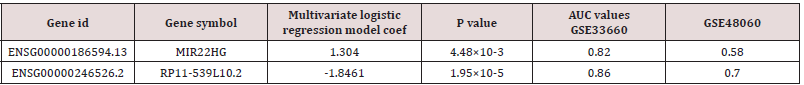
By subjecting the lnc RNAs expression data to multivariate logistic regression model, we found two lnc RNAs were significantly different expression in the MI (multivariate p<0.05, Table 2). We then generate a distinguish score with the two lnc RANs (RP11- 539L10.2 and MIR22HG) to distinguish MIs from controls. The distinguish score formula was developed according to the expression levels of two lnc RNAs as follows: distinguish score = (-1.8461 × expression level of RP11-539L10.2) + (1.304 × expression level of MIR22HG) (Table 3). The distribution of the lncRNA distinguish score and expression signature were shown in Figure 2. We found that patients with high- distinguish scores tended to express high levels of MIR22HG and low level of RP11- 539L10.2 in their circulating cells. In addition, receiver operating characteristic curve (ROC) analysis was conducted to assess the predictive accuracy of the two-lnc RNAs signature (Figure 3). The results showed a prognostic power of distinguishing AMIs from controls either in the training dataset (AUC=0.92). Further, we calculated the AUCs for each of the lncRNA and found the two lnc RNAs showed a good distinguish performance (AUC>0.5; Table 3).
Figure 2: LncRNA risk score analysis.
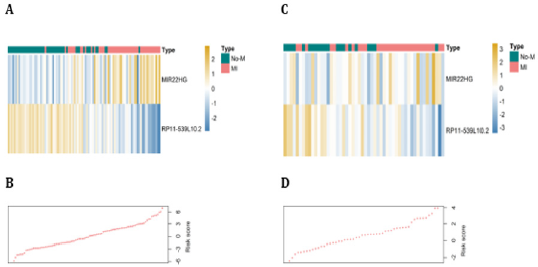
The distribution of 2-lncRNA risk score and lncRNA expression signature were analyzed in the train and validate datasets. Heatmap of the lncRNA expression profiles in the train (A) and validate (C) datasets. Rows and columns represent lnc RNAs and patients, respectively. (A) lncRNA signature risk score distribution in the training (B) and validating (D) datasets.
Figure 3: ROC curves assess the accuracy of our defined signature.
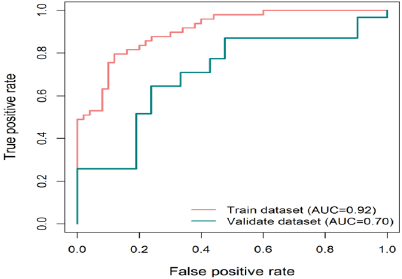
ROC curves assess the accuracy of the lncRNA signature in the train (red line) and validate (green line) datasets. True positive rate represents sensitivity, whereas false positive rate is one minus the specificity.
Functional Annotation
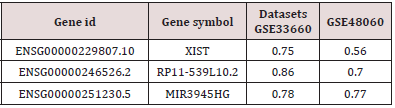
To infer the potential biological function of the lnc RNAs, the co-expressed relationships between the expression levels of six lnc RNAs and protein-coding genes (PCGs) were tested in both datasets. Based on the criterion of Person correlation coefficients higher than 0.4, MIR3945HG was shown to be co-expressed with 303 PCGs (Table 2). GO function enrichment analysis for these PCGs was then performed, suggested that these PCGs were significantly enriched in 7 GO terms (Figure 4A), which clustered in inflammatory response, innate immune response, regulation of cytokines secretion, leukocyte migration, immune response, MyD88-dependent toll-like receptor signaling pathway, and defense response to bacterium and chemotaxis (Figure 4, Table 3). In general, function annotation results indicated that MIR3945HG might participate in the development of AMI through interacting with immune response related PCGs.
Figure 4: GO enrichment analysis for the function of MIR3945HG.
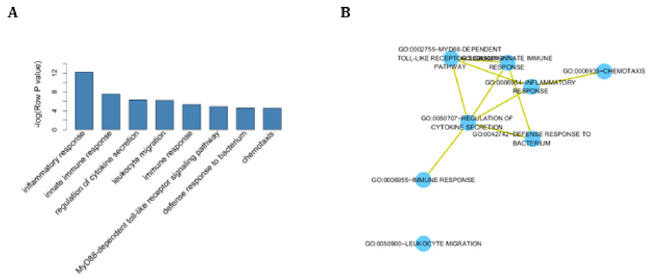
(A) The original significance outputted from DAVID for GO biological processes were transformed in to ‘–log (P-value)’ for plotting. (B) The functional map of enriched GO terms with each node indicates an enriched GO term and each edge represents the common genes shared between connecting enriched GO.
Discussion
The study of gene biology functions has mainly focus in protein-coding genes and miRNAs until the discovery of thousands of functional regulatory lnc RNAs [27]. Lnc RNAs, show great tissue- and disease-specific expression levels compared with protein-coding genes, are dysregulated in many disease types, which are believed to play important roles in regulating several biological processes. It has been reported that dysregulation in lnc RNAs expression is associated with some cardiovascular diseases risk [28]. The functions of lncRNA in AMI have not been fully understood [29,30]. In our study, by data mining two previously published gene expression microarray data from GEO, we obtained the expression pattern of lnc RNAs in AMI patients and control subjects. Six lnc RNAs were identified to be associated with AMI, and a set of two-lncRNA (MIR22HG and RP11-539L10.2) was signature shown power in distinguishing AMI from controls either in the training or in the validation series. Although the number of lnc RNAs discovered and recorded in biological databases, such as Ensemble [31] and GENCODE [30] is increasing, only a few lnc RNAs were fully functionally characterized. In our study, we found that six lnc RANs (MIR22HG, RP11-296O14.3, IDI2-AS1, RP11- 539L10.2, MIR3945HG, RP11-96D1.11) were associated with AMI. Voellenkle C et al. reported differential expression of MIR22HG in human umbilical vein endothelial cells between normoxia and hypoxia status, and they validated this finding in a mouse model of hindlimb ischemia, suggesting an important role of MIR22HG in the vascular physiopathology [30].
MIR3945HG is also observed to be aberrantly expressed in lung squamous cell carcinoma, and its high expression is associated with longer survival time of lung squamous cell carcinoma patients [31]. To gain a deeper knowledge of the above mentioned lnc RNAs in AMI, the underlying regulatory mechanisms should be further studied (Table 4).
Study limitations
First, owing to the restricted availability of data, a fraction (about 3800 in more than 20 thousand) but not all of the human lnc RNAs were included in our analysis. Second, we tested the associations between the expression of lnc RNAs and AMI and identified some candidate lnc RNAs might participate in the development of AMI, but the mechanisms are not clear. The functions of lnc RNAs identified in this study need to be explored in further experimental study. Third, the lnc RNAs expression from circulating endothelial cells in the training dataset and from circulating nucleated cells of the blood in the validating dataset. This is also showed that robustness of the three-lncRNA signature in circulating cells is robust in distinguishing AMI form controls.
Conclusion
Our study presents a two-lnc RNAs signature significantly related with AMI. This signature might contribute to diagnose AMI patients. Changes of lnc RNAs’ expression level in the circulating cells may reflect the underlying biological mechanisms for AMI under detection. Our findings suggest that expression profiling of the lncRNA complement of the cardiac transcriptome in the systemic circulation might provide new approach for early diagnosis and treatment of the AMI.
References
- Suresh R, Li X, Chiriac A, Goel K, Terzic A, et al. (2014) Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. Journal of Molicular Cell Cardiology 74:13-21.
- Townsend N, Wilson L, Bhatnagar P, Wickramasinghe K, Rayner M, et al. (2016) Cardiovascular disease in Europe: epidemiological update 2016. European Heart Journal 37(42): 3232-3245.
- Gaziano T A GJM (2016) Myocardial Infarction: A Companion to Braunwald's Heart Disease E-Book 2016.
- Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, et al. (2013) Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation 127(1): e6-e245.
- Antman EM, Cohen M, Bernink PJ, McCabe CH, Horacek T, et al. (2000) The TIMI risk score for unstable angina/non-ST elevation MI: A method for prognostication and therapeutic decision making. JAMA 284(7): 835-842.
- Granger CB, Goldberg RJ, Dabbous O, Pieper KS, Eagle KA, et al. (2003) Predictors of hospital mortality in the global registry of acute coronary events. Arch International Medical 163(19): 2345-2353.
- Law MR, Watt HC, Wald NJ (2002) The underlying risk of death after myocardial infarction in the absence of treatment. Arch Intern Med 162(21): 2405-2410.
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, et al. (2007) Genomewide association analysis of coronary artery disease. National England Journal Med (5): 443-453.
- Peters T, Scheren B (2014) Missing links in cardiology: long non-coding RNAs enter the arena. Pflugers Arch 466(6): 1177-1187.
- Lipovich L, Johnson R, Lin CY (2010) MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta 1799(9): 597-615.
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, et al. (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152(3): 570-583.
- Yang KC, Yamada KA, Patel AY, Topkara VK, George I, et al. (2014) Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129(9): 1009-1021.
- Holdt LM, Beutner F, Scholz M, Gielen S, Gäbel G, et al. (2010) ANRIL Expression Is Associated With Atherosclerosis Risk at Chromosome 9p21. Arteriosclerosis, Thrombosis, and Vascular Biology 30(3): 620-627.
- Tsai PC, Liao YC, Lin TH, Hsi E, Yang YH, et al. (2012) Additive effect of ANRIL and BRAP polymorphisms on ankle-brachial index in a Taiwanese population. Circ Journal 76(2): 446-452.
- Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, et al. (2012) Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 220(2): 4494-55.
- Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, et al. (2015) Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Europian Heart Journal 36(6): 353-368a.
- Zhai H, Li XM, Liu F, Chen BD, Zheng H, et al. (2017) Expression pattern of genome-scale long noncoding RNA following acute myocardial infarction in Chinese Uyghur patients. Oncotarget 8(19): 31449-31464.
- Zhang Y, Sun L, Xuan L, Pan Z, Li K, et al. (2016) Reciprocal Changes of Circulating Long Non-Coding RNAs ZFAS1 and CDR1AS Predict Acute Myocardial Infarction. Science Rep 6: 22384.
- Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, et al. (2013) Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biology 20(7): 908-913
- Du Z, RGW Fei T Fau Verhaak, Verhaak Rg Fau Su Z, Su Z Fau hang Y, Zhang Y Fau Brown M, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer Nat Struct Mol Biology 20(7): 908-913.
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Research 22(9): 1760-1774.
- Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, et al. (2011) Strategies for aggregating gene expression data: the collapse Rows R function. BMC Bioinformatics 12: 322.
- Team RC (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing Austria.
- Liao Q, Liu C, Yuan X, Kang S, Miao R, et al. (2018) Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res 39(9): 3864-3878.
- Huang DW, Sherman BT, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. National Protocols 4(1): 44-57.
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5(11): e13984.
- Guttman M, Amit I, Garber M, French C, Lin MF, et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235): 223-227.
- Uchida S, Dimmeler S (2015) Long Noncoding RNAs in Cardiovascular Diseases. Circulation Research 116(4): 737.
- Lazarevic V, Margot P, Soldo B, Karamata D (1992) Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. Journal of General Microbiology 138(9): 1949-1961.
- Chen WJ, Tang RX, He RQ, Li DY, Liang L, et al. (2017) Clinical roles of the aberrantly expressed lncRNAs in lung squamous cell carcinoma: a study based on RNA-sequencing and microarray data mining. Oncotarget 8(37): 61282-61304.
- Voellenkle C, Garcia Manteiga JM, Pedrotti S, Perfetti A, De Toma I, et al. (2016) Implication of long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Scientific reports 6: 24141.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...