
Lupine Publishers Group
Lupine Publishers
Menu
ISSN: 2637-4749
Research Article(ISSN: 2637-4749) 
Association between the Presence of Intrauterine Escherichia coli Virulence Genes and Subsequent Reproductive Tract Disease in Postpartum Dairy Cows Volume 2 - Issue 3
José Denis-Robichaud, John Morris Fairbrother, Flavien Ndongo Kassé and Jocelyn Dubuc*
- Université de Montréal, Faculté de médecine Vétérinaire, St-Hyacinthe, Québec, Canada
Received: March 14, 2019; Published: March 25, 2019
Corresponding author: Jocelyn Dubuc, Faculté de médecine vétérinaire, Université de Montréal, rue Sicotte, St-Hyacinthe, Québec, Canada
DOI: 10.32474/CDVS.2019.02.000140
Abstract
An association between postpartum intrauterine Escherichia coli and subsequent reproductive tract diseases such as purulent vaginal discharge (PVD) and endometritis (ENDO) has been found inconsistently in previous research. This inconsistency may be due to differences in the pathogenicity and presence of certain virulence factors in the various strains. The objective of this study was to evaluate the association between the presence of intrauterine E. coli virulence factor (VF) genes after parturition and subsequent reproductive tract diseases in postpartum dairy cows. Intrauterine swabs were collected from cows 4 (± 3) DIM. The swabs were plated to identify E. coli, Trueperella pyogenes, Fusobacterium necrophorum, and Prevotella melaninogenica. A subgroup of the E. coli samples was submitted for colony hybridization for identification of 40 VF genes. Purulent vaginal discharge and ENDO were diagnosed at 35 (±7) DIM using the Metricheck device (purulent discharge or worse) and the cytobrush technique adapted for use in cattle (≥ 6% polymorphonuclear leukocytes), respectively. Cows diagnosed with PVD, ENDO, or both conditions were classified as positive for reproductive tract disease. Logistic regression models were built using the reproductive tract disease status as the outcome, and the bacteria and VF gene presence as the exposure. Of the 465 cows enrolled, 52% of the uterine samples were positive for E. coli, 34% were positive for T. pyogenes, 3% were positive for F. necrophorum, and 1% were positive for P. melaninogenica. A total of 152 E. coli samples were examined for VF gene identification. Reproductive tract disease was diagnosed in 237 cows (51%). The presence of intrauterine E. coli and T. pyogenes was associated with greater odds of reproductive tract disease. Cows with E. coli positive for VF genes fepC, maIX, hlyE, sitA, irp1, irp2, fyuA, or iss had greater odds of having subsequent reproductive tract disease compared to cows without E. coli. These VF genes code for iron acquisition, the maltose and glucose PTS system, hemolysin E toxin, and increased serum survival. Three of the siderophore genes (irp1, irp2, and fyuA) are part of the core of a high-pathogenicity islands, previously described in extraintestinal pathogenic E. coli (ExPEC) The results of this study suggest that certain VFs are likely to contribute to the pathogenicity of E. coli strains as they are associated with subsequent reproductive tract disease.
Keywords:Endometritis; High-Pathogenicity Island; Purulent Vaginal Discharge; Siderophore
Introduction
Purulent vaginal discharge (PVD) and endometritis (ENDO) are reproductive tract diseases diagnosed in postpartum dairy cows and are associated with a detrimental impact on subsequent reproductive performance Gilbert et al. [1] Dubuc et al. [2], Denis- Robichaud and Dubuc [3]. The diagnosis of PVD is based on the presence of purulent material in the vagina visualized using devices such as the Metricheck or a vaginoscope LeBlanc et al. [4], McDougall et al. [5], Barlund et al. [6]. The diagnosis of ENDO is based on the presence of an excessive proportion of inflammatory cells on an endometrial smear obtained with a cytobrush or a lowvolume uterine lavage Wagener et al. [7]. Risk factors for PVD and ENDO are dystocia, retained placenta, postpartum metritis, as well as negative energy balance Leblanc et al. [4], Hammon [8], Dubuc et al. [9], suggesting that both the immunity of the cow and the presence of intrauterine bacteria shortly after parturition play a role in the later development of these reproductive diseases. However, intrauterine bacteria such as Escherichia coli, Trueperella pyogenes, Fusobacterium necrophorum, and Prevotella melaninogenica are associated inconsistently with endometrial inflammation and clinical disease Williams et al. [10], Wagener et al. [11], Bicalho et al. [12]. Research suggests that the association between the presence of different bacteria and reproductive tract disease depends on the moment of sampling, due to the dynamic nature of the infections Bicalho et al. [13], Prunner et al. [14], Wagener et al. [11]. Whereas the presence of intrauterine T. pyogenes during the first month postpartum was associated with PVD and endometritis, the association between E. coli during the first month postpartum and these diseases is not as clear Bicalho et al. [13], Prunner et al. [14], Wagener et al. [11].
A variety of strains of E. coli with differing pathogenicity have been found, possibly explaining the inconsistent association between the intrauterine presence of this bacterium and the subsequent development of reproductive tract disease Kaper et al. [15], Sheldon et al. [16]. Intrauterine E. coli has been characterized by the identification of virulence factor (VF) genes Silva et al. [17], Bicalho et al. [13], Kassé, et al. [18]. Bicalho, et al. [13] found an association between the presence of certain VF genes (fimH, cdt and astA) and PVD. Dubuc, et al. [9] showed however, that cows without PVD can have endometritis, and vice versa, which could bias the associations between risk factors and disease toward the null hypothesis if a proportion of the cows classified as healthy also have reproductive tract disease Dohoo et al. [19]. The objective of this study was to evaluate the association between the presence of intrauterine E. coli, and E. coli VF genes after parturition and subsequent reproductive tract disease (PVD, ENDO, or both) in postpartum dairy cows.
Materials and Methods
This study was originally designed to identify associations between E. coli VF genes and postpartum metritis (PPM), as reported by Kassé et al. [18]. In this work, Holstein dairy cows from 4 commercial farms located within 30 km of the bovine ambulatory clinic of the Faculté de médecine vétérinaire, Université de Montréal (Saint-Hyacinthe, QC, Canada) were enrolled on a prospective cohort study from November 2011 to June 2012. Farms were a convenient sample of freestall housed herds using computerized health records, and enrolled in a biweekly herd health veterinary program. During the study period, all cows that did not yet show clinical signs of PPM were enrolled weekly (at 4 ± 3 DIM). The sample size was calculated for the original study Kassé et al. [18], to identify a 20% difference in PPM prevalence with 95% confidence and 80% power Dohoo et al. [19]. Procedures were approved by the animal care committee of the Université de Montréal [11-Rech-1605]. At enrolment, cows were sampled to identify uterine bacteria. Briefly, cows were restrained and the perineum was cleaned and disinfected with 70% ethyl alcohol solution (Isopropylic Alcohol 70% USP; Green Field Inc., Brampton, ON, Canada). A sterile double-guarded uterine swab (Guarded culture swab; Jorvet Inc., Loveland, CO, US) was introduced in the cranial vagina then passed through the cervix until the body of the uterus. The swab was then exposed to the dorsal aspect of the uterine wall. The swab was placed in an anaerobic transportation medium (BBL Port-A-Cult Tubes; Becton, Dickinson and Company, Sparks, MD, US) and kept at 4°C until submission to the veterinary diagnostic laboratory of the Université de Montréal within 12h of collection.
Escherichia coli, T. pyogenes, F. necrophorum, and P. melaninogenica were identified from the uterine swab in the veterinary diagnostic laboratory of the Université de Montréal (Saint-Hyacinthe, QC, Canada), as described by Kassé et al. [18]. Briefly, swabs were plated on blood agar and MacConkey agar (Oxoid) at 37°C for isolation of E. coli. Five isolates from each positive sample were submitted to indole spot, Simmons citrate, and motility tests, for confirmation of E. coli. Escherichia coli isolates were stored in tryptic soy broth containing 30% glycerol at −80°C (Becton, Dickinson and Company) for further analysis. Swabs were also plated on Colombia blood agar (Oxoid, Ottawa, ON, Canada) at 35°C for 48 h for isolation of T. pyogenes using the PON- BAC-019 procedure (beta-hemolytic, catalase-negative minuscule colonies demonstrating gram-positive coryneform rods; McVey et al., 2013). A subgroup of the E. coli isolates was examined by colony hybridization using radioactively labeled (32P) DNA probes for identification of VF genes at the World Organisation for Animal Health Reference Laboratory for Escherichia coli (Faculté de médecine vétérinaire, Université de Montréal; Maluta et al. [20], Fairbrother et al. [21]. Briefly, the isolates were spotted onto Luria- Bertani agar and incubated at 37°C overnight. Colonies were then transferred to Whatman 541 filter paper (Whatman, Piscataway, NJ, US). The filter papers were processed, hybridized, and visualized by autoradiography. Probes were derived from E. coli control strains by uniplex PCR, using the primers of the tested genes. After amplification, PCR products were purified and concentrated, using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. After purification, probes were marked with phosphate 32 using a specific kit (Amersham Ready to go DNA Labeling Beads, GE Healthcare UK Limited, Little Chalfont, UK), according to the manufacturer’s instructions. All isolates (5 per sample) submitted for colony hybridization were tested for 40 VF genes (complete list in Appendix A1).
Postpartum metritis was diagnosed and recorded between calving and 21 DIM by one person per farm. The standardized definition was reviewed with participating farmers before the beginning and every 3 mo during the study: fetid watery red-brown uterine discharge, associated with fever (rectal temperature > 39.5°C), and systemic signs of illness (dullness, reduced appetite, and milk production; Drillich et al. [11]. When PPM was diagnosed, cows were treated with 5 d of ceftiofur IM SID (2.2 mg/kg; Zoetis Animal Health, Kirkland, QC, Canada). Purulent vaginal discharge and ENDO were diagnosed biweekly at 35 (±7) DIM. Firstly, the vaginal discharge score was assessed using the Metricheck device (0 = no discharge, 1 = clear mucus, 2 = mucus with flecks of pus, 3 = mucopurulent discharge, 4 = purulent discharge or 5 = foul smelling discharge; McDougall et al. [5]. Then, an endometrial cytology sample was taken using the cytobrush technique adapted for use in cattle Kasimanickam et al. [22]; Dubuc et al. [9]. Immediately after collection, the cytobrush was rolled on a microscope glass slide to obtain a smear. The microscope slides were stained within 12 h of collection with a modified Wright- Giemsa stain (Hema3; Biochemical Sciences, Swedesboro, NJ) and glass coverslips were applied when dry as previously described Dubuc et al. [9]. The cytology slides were used to determine the percentage of polymorphonuclear leukocytes (PMNL) among two hundred cells (PMNL and endometrial cells) by 2 observers. Slide readers were blinded to on-farm findings and treatment allocation. Purulent vaginal discharge was defined as a vaginal discharge score of ≥ 4, and ENDO was defined as percentage of PMNL ≥ 6% Denis- Robichaud and Dubuc [3]. Cows diagnosed with PVD, ENDO, or both were classified as positive for reproductive tract disease.
Statistical Analyses
All analyses were performed using SAS Studio 3.6 (SAS Institute Inc., Cary, NC, US), the cow being the experimental unit. Parity (1st, 2nd, 3rd and greater) and season of calving (winter: November to February, spring: March to June) were extracted from the computerized record system (DSAHR Inc., Saint-Hyacinthe, QC, Canada). Frequencies were calculated (PROC FREQ) for binary and categorical variables. Based on the bacteriological culture results, cows were categorized as (0) no bacteria, (1) presence of E. coli combined with other bacteria (T. pyogenes, F. necrophorum, or P. melaninogenica), (2) presence of a E. coli alone, or (3) presence of other bacteria (T. pyogenes, F. necrophorum, or P. melaninogenica) only. The same categories (0 to 3) were also created for T. pyogenes, F. necrophorum, and P. melaninogenica. Based on the bacteriological culture and the colony hybridization results, cows were also classified as (0) negative for E. coli, (1) positive for E. coli but negative for the VF gene, or (2) positive for E. coli and for the VF gene. Logistic regression models (PROC GLIMMIX) were built using the reproductive tract disease status as the outcome to assess its association with the presence of different intrauterine bacteria, and with the presence of E. coli VF genes. Herd was included in all models as a fixed effect for accounting for clustering, and parity and season were included as confounders if their P-value was > 0.20 (Maldonado and Greenland, 1993). Models were presented if statistical significance (P < 0.05) or tendency to significance (0.05 ≤ P < 0.10) was reached. The difference between parameters of a categorical variable was assessed using Tukey-Kramer adjustment for multiple comparisons (PTK), using < 0.05 for statistical significance and 0.05 to 0.10 for tendency to significance. The Hosmer and Lemeshow goodness-of-fit test was used to assess the fit of the models. Outlier (Pearson and deviance residuals), extreme (hat matrix), and influential (DFBeta) covariate patterns were assessed, and models were tested without extreme and influential values to ensure robustness of the coefficients Dohoo et al. [19].
Results
A total of 486 cows were enrolled in this study from 4 freestall commercial farms (141, 177, 136, and 32 samples from farms 1, 2, 3, and 4, respectively). A total 465 cows remained for final analyses as 21 (4%) were culled before the diagnosis of reproductive tract diseases at 35 DIM. Thirty-four percent (n = 160), 32% (n = 148), and 34% (n = 157) of the remaining cows were of 1st, 2nd, and 3rd and greater parity, respectively. Forty-two percent (n = 194) of the cows calved during spring, and 58% (n = 271) calved during winter. Overall, 243 (52%) of the uterine samples were positive for E. coli, 156 (34%) were positive for T. pyogenes, 16 (3%) were positive for F. necrophorum, and 3 (1%) were positive for P. melaninogenica. A total of 780 isolates from 152 of the samples positive for E. coli were submitted for hybridization to identify VF genes. A visual summary of the 36 VF genes found in the uterine E. coli of cows is presented in Figure 1. A total of 133 different cow VF profiles were identified, with 1 to 16 VF per profile. The most prevalent VF genes were hlyE (n = 135; 89% cows harboring one or more positive isolates) and fimH (n = 133; 88% cows harboring one or more positive isolates), coding for hemolysin and adherence, respectively. Postpartum metritis was diagnosed in 65 cows (14%), and reproductive tract diseases were diagnosed in 237 cows (51%), of which 32 (7%) had PVD, 136 (29%) had endometritis, and 69 (15%) had both PVD and endometritis.
Denis-Robichaud, Figure 1.
Figure 1: Presence of 36 virulence factor genes in intrauterine Escherichia coli isolates from 152 Holstein dairy cows sampled 4±3 DIM. Cows’ profiles are sorted by reproductive tract disease status at 35±7 DIM (vaginal purulent discharge: PVD, endometritis: ENDO).
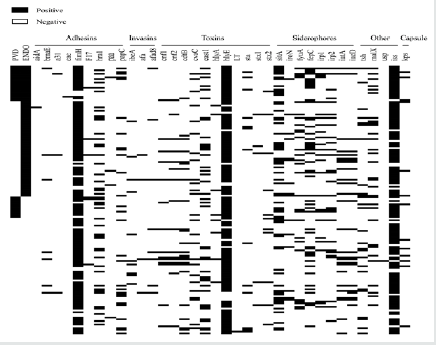
Table 1 presents the odds of reproductive tract diseases in cows without intrauterine bacteria, and with different profiles of E. coli, T. pyogenes, F. necrophorum, and P. melaninogenica. Virulence factor genes fepC, maIX, hlyE, and sitA were associated with a harmful effect and VF gene cdtB with a protective effect for reproductive tract disease (Figure 2). Cows harboring isolates positive for fepC had 2.62 times the odds of having reproductive disease compared to cows without E. coli (95% CI = 1.26-5.45; PTK = 0.01), and 2.09 times the odds of having reproductive disease compared to cows with E. coli but without the fepC (95% CI = 0.95-4.62; PTK = 0.07). Cows harboring isolates positive for maIX had 2.99 times the odds of having reproductive disease compared to cows without E. coli (95% CI = 1.20-7.43; PTK = 0.02), and 2.25 times the odds of having reproductive tract disease compared to cows with E. coli but without the maIX (95% CI = 0.87-5.80; PTK = 0.09). Cows harboring isolates positive for hlyE and sitA had 1.72 (95% CI = 1.10-2.69; PTK = 0.04), and 2.13 (95% CI = 1.14-3.97; PTK = 0.02) times the odds of having reproductive tract disease compared to cows without E. coli, respectively. Cows with E. coli but without VF gene cdtB were 1.80 times more likely to have reproductive tract disease than cows without E. coli (95% CI = 1.14-2.83; PTK = 0.01), and cows harboring E. coli positive for cdtB had 0.41 times the odds of having reproductive tract disease compared to cows with E. coli and without cdtB (95% CI = 0.16-1.05; PTK = 0.06). Logistic regression models for irp1, irp2, fyuA, and iss had a tendency to be statistically significant, but there was a significant difference in the odds of reproductive tract disease between cows harboring E. coli positive for these VF genes and cows without E. coli (PTK < 0.05). Cows harboring E. coli positive for irp1, irp2, fyuA, and iss had 3.62 (95% CI = 1.10-11.90; PTK = 0.03), 2.51 (95% CI = 1.01-6.24; PTK = 0.05), 2.69 (95% CI = 1.01-7.18; PTK = 0.05), and 1.69 (95% CI = 1.08-2.64; PTK = 0.02) times the odds of having reproductive tract diseases compared to cows without E. coli, respectively.
Denis-Robichaud, Figure 2.
Figure 2: Proportion of cows with reproductive tract disease (purulent vaginal discharge, endometritis, or both) at 35 ± 7 DIM stratified by uterine bacteriological status at 4±3 DIM in 374 Holstein dairy cows. The marginal means (± SEM) were obtained from logistic regression models (one per virulence factor gene), adjusted for herd clustering using a Tukey-Kramer adjustment for multiple comparisons. The virulence factor genes fepC, maIX, hlyE, and sitA were identified as harmful, and the virulence factor gene cdtB was identified as protective for reproductive diseases. * PTK < 0.05. † PTK < 0.10.
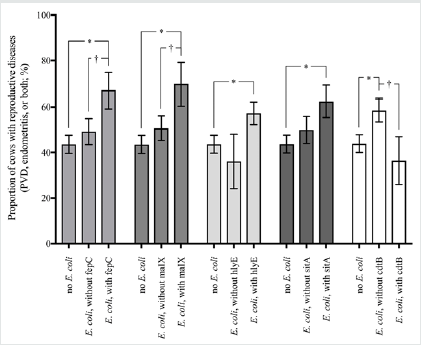
Table 1: Odds of reproductive tract disease at 35±7 DIM (purulent vaginal discharge, endometritis, or both) from logistic regression models in 465 Holstein dairy cows with different profiles of uterine bacterial status at 4±3 DIM, adjusted for herd clustering.
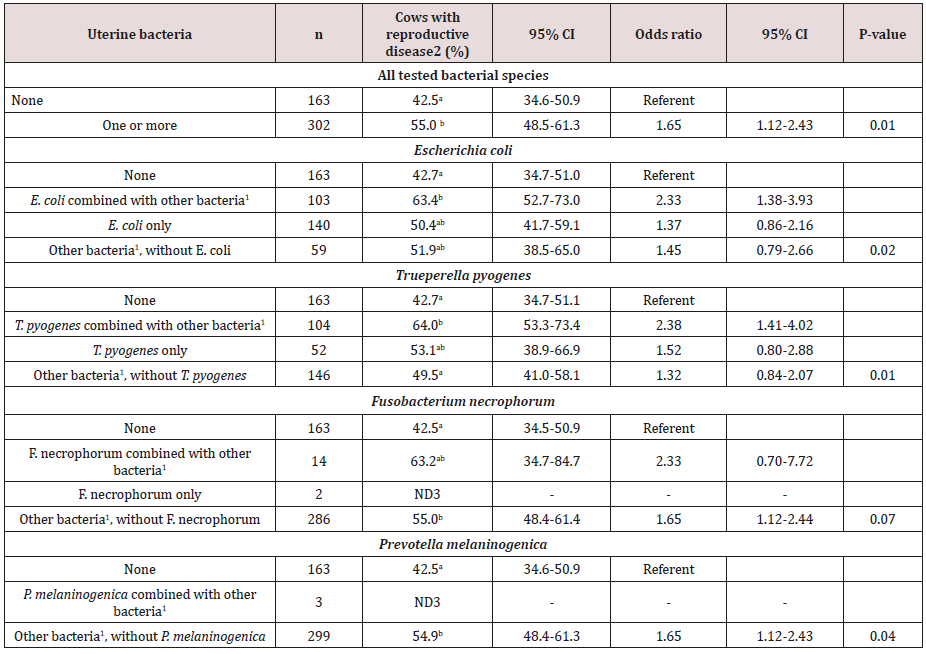
a-bMeans within a column within a model with different subscripts differ, assessed using Tukey-Kramer adjustment for multiple comparisons (PTK < 0.05).
1Other bacterial species were Escherichia coli, Trueperella pyogenes, Fusobacterium necrophorum, and Prevotella melaninogenica
2Estimated population marginal proportion (LSM) of cows with reproductive diseases at 35±7 DIM (purulent vaginal discharge, endometritis, or both)
3ND: not determined due to small n
Discussion
As the presence of postpartum intrauterine E. coli was inconsistently associated with reproductive tract diseases in dairy cows Bicalho et al. [13], Prunner et al. [14], Wagener et al. [11], the objective of the present study was to assess the association of reproductive tract diseases such as PVD and ENDO with the presence of intrauterine bacteria during the first week postpartum of dairy cows, and more specifically the presence of E. coli VF genes. When compared to cows without intrauterine E. coli, cows with genes for iron acquisition (fepC, sitA, irp1, irp2, and fyuA), for maltose and glucose PTS system (malX), for hemolysin E toxin (hlyE), and for increased serum survival (iss) had greater odds of presenting reproductive disease.
As observed for urinary infection, meningitis, and septicemia in humans, the E. coli involved in reproductive tract disease require characteristics allowing them to survive outside of the intestinal environment. Iron acquisition factors are often associated with extraintestinal pathogenic E. coli (ExPEC) as they contribute to the survival of the bacteria in an environment where iron is not readily available Litwin and Calderwood [23]. Moreover, it was suggested that siderophores also had a cytotoxic effect on immune cells Autenrieth et al. [24], Schubert et al. [25]. In the present study, five siderophore genes were associated with increased odds of reproductive tract diseases, of which three (irp1, irp2, and fyuA) are part of the core of the high-pathogenicity islands (HPI). First described in pathogenic Yersinia, HPI results from an accumulation of virulence factors on the bacterial chromosome and have been identified in pathogenic ExPEC, suggesting that they contribute to the survival and pathogenicity of bacteria Schubert et al. [25], Dezfulian et al. [26], Tourret et al. [27]. Septicemia and pelvic inflammatory disease in cattle have been associated with E. coli positive for the HPI and fyuA genes, respectively Dezfulian et al. [26], Sheldon, et al. [16], but the present study was the first to demonstrate an association between reproductive tract disease and fyuA, irp1 and irp2 genes.
In the present study, both malX and iss were also associated with increased odds of reproductive tract disease. These two factors play a role the survival of ExPEC outside of the intestinal environment. malX is mainly considered as a marker for pathogenicity as its codes for an enzyme system that does not contribute to the virulence of a bacteria but has been associated with other virulence genes Johnson et al. [28,29], Östblom, et al. [30]. It was, however, found in persistent E. coli strains which suggested that the malX gene may also have an additive or synergetic effect with respect to pathogenicity islands Östblom, et al. [30]. Similarly, iss contributes to the survival of the bacteria against host immunity affecting the complement system Lynne et al. [31] but was mainly associated with other virulence factors Chuba et al. [32], Johnson [33]. Our finding of an association between reproductive tract disease and pathogenicity markers such as malX and iss suggests that the impact of intrauterine E. coli is influenced by the VFs of the bacteria, possibly explaining the inconsistent results previously found Williams et al. [10], Wagener, et al. [11], Bicalho et al. [12]. The eight factors associated with reproductive tract diseases in the present study were different from those found previously by Bicalho et al. [12]; fimH, cdt, and astA). Whereas Bicalho et al. [13] examined the association between the VFs and PVD, reproductive tract diseases in the present study were defined as either PVD, ENDO, or both. In studies looking at both PVD and ENDO, 10 to 26% of the cows were negative for PVD and positive for ENDO Dubuc et al. [2], Denis- Robichaud and Dubuc [3], which could have contributed to the different results between studies. It is not clear why, in the present study, the presence of E. coli harboring the cdtB gene was associated with lower odds of reproductive disease than the presence of E. coli without this gene or the absence of E. coli. A possible explanation is that the analyses did not account for a confounder when assessing the association between the presence of a gene and reproductive disease Dohoo et al. [19]. For example, if the presence of cdtB gene was associated with an unmeasured variable that reduces the odds of reproductive disease, the protective effect of this gene would be the eventual consequence.
In the present study, bacterial culture was used to identify four bacterial species (E. coli, T. pyogenes, F. necrophorum, and P. melaninogenica). The prevalence of cows with intrauterine bacteria of these species was lower than in other studies looking at the presence of intrauterine bacteria where more or different bacterial species were identified Griffin et al. [34], Sheldon, et al. [35], Wagener et al. [11]. Wagener et al. [11] found a prevalence of intrauterine bacteria of 80% at 3 DIM, although Streptococcus uberis, which we did not look for, was present alone in 18% of the cows. In the present study, it is possible that the association between the presence of intrauterine bacteria and reproductive tract disease was biased toward the null hypothesis due to the presence of other bacterial species in the cows we classified as negative to intrauterine bacteria. For example, the presence of S. uberis at 3 DIM was associated with greater odds of PVD Wagener et al. [11]. Similarly, metagenomic analysis has the potential to identify uncultivable bacteria Santos et al. [36], which could give rise to different findings with respect to the association between the presence of intrauterine bacteria and reproductive tract diseases. The prevalence of reproductive disease in the cows classified as negative for intrauterine bacteria was greater than 40% in our study, which could partly be explained by the limited number of bacterial species that we looked for, as well as the point in time that the uterine sample was taken. Recent studies showed the dynamic nature of the intrauterine bacterial population Bicalho et al. [13], Prunner et al. [14], Wagener, et al. [11], which was not evaluated in the present study as only one sample was taken shortly after calving. Finally, the prevalence of reproductive tract disease in cows without intrauterine bacteria was 42.5%, which supports the idea that even though bacteria and their VFs are playing a role in the development of the disease, other factors are likely involved Hammon et al. [8], Sheldon et al. [37], Dubuc, et al. [9], [38-45].
Conclusion
The essential point of the life cycle is that whereas one nematode egg can develop into only one adult, one trematode egg may eventually develop into hundreds of adults. This is due to the phenomenon of paedogenesis in the molluscan intermediate host, i.e. the production of new individuals by single larval forms. The adult flukes are always oviparous and lay eggs with an operculum or lid at one pole. In the egg, the embryo develops into a pyriform (pear-shaped), ciliated larva called a miracidium. The miracidium, propelled through the water by its cilia, does not feed and must, for its further development, find a suitable snail within a few hours. It is believed to use chemotactic responses to ‘home’ on the snail and, on contact, it adheres by suction to the snail and penetrates its soft tissues aided by a cytolytic enzyme. The entire process of penetration takes about 30 minutes after which the cilia are lost and the miracidium develops into an elongated sac, the sporocyst, containing a number of germinal cells. These cells develop into rediae which migrate to the hepato-pancrcas of the snail; rediae are also larval forms possessing an oral sucker, some flame cells and a simple gut. From the germinal cells of the rediae arise the final stages, the cereariae, although if environmental conditions for the snail are unsuitable, a second or daughter generation of rediae is often produced instead. The cercaria, young flukes with long tail emerge actively from the snail usually in considerable number. Once a snail is infected, cercariae continue to be produced indefinitely although the majority of infected snails die prematurely from gross destruction of the hepatopancreas. Typically the cercariae swim for some time, utilizing even a film of water, and within an hour or so attach themselves to vegetation, shed their tails and encyst. This stage is called a metacercaria. EncyThis study showed that the presence of intrauterine E. coli with genes for siderophores or HPI during the first week postpartum in dairy cows was associated with greater odds of reproductive tract disease [45-50]. Some of these genes have previously been shown to co-exist in ExPEC and possibly contribute to the survival of E. coli in the reproductive tract [51-56]. These findings support the idea that peripartum bacterial contamination has an impact on subsequent health of the reproductive tract, and that it is possible to identify more accurately the bacteria involved using VFs [56-62].
Acknowledgement
The data used in the present study were retrieved from a research project funded by “Le Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec– Programme Innov’Action” (Québec, QC, Canada) with a research grant attributed to Jocelyn Dubuc (810144). The authors are grateful to Brigitte Lehoux, Ghyslaine Vanier, and Jean-Philippe Pelletier (Université de Montréal, St- Hyacinthe, QC, Canada) for their technical work during this project and to the participating dairy farmers for their willingness to enroll in this study.
References
- Gilbert RO, ST Shin, CL Guard, HN Erb, M Frajblat (2005) Prevalence of endometritis and its effects on reproductive performance of dairy cows. Theriogenology 64: 1879-1888.
- Dubuc J, TF Duffield, KE Leslie, JS Walton, SJ LeBlanc (2010) Definitions and diagnosis of postpartum endometritis in dairy cows. J Dairy Sci 93: 5225-5233.
- Denis Robichaud J, J Dubuc (2015) Determination of optimal diagnostic criteria for purulent vaginal discharge and cytological endometritis in dairy cows. J Dairy Sci 98: 6848-6855.
- LeBlanc SJ, TF Duffield, KE Leslie, KG Bateman, GP Keefe, et al. (2002) Defining and diagnosing postpartum clinical endometritis and its impact on reproductive performance in dairy cows. J Dairy Sci 85: 2223-2236.
- McDougall S, R Macaulay, C Compton (2007) Association between endometritis diagnosis using a novel intravaginal device and reproductive performance in dairy cattle. Anim Reprod Sci 99: 9-23.
- Barlund CS, TD Carruthers, CL Waldner, CW Palmer (2008) A comparison of diagnostic techniques for postpartum endometritis in dairy cattle. Theriogenology 69: 714-723.
- Wagener K, C Gabler, M Drillich (2017) A review of the ongoing discussion about definition, diagnosis and pathomechanism of subclinical endometritis in dairy cows. Theriogenology 94: 21-30.
- Hammon DS, IM Evjen, TR Dhiman, JP Goff, JL Walters (2006) Neutrophil function and energy status in Holstein cows with uterine health disorders. Vet. Immunol. Immunopathol 113: 21-29.
- Dubuc J, TF Duffield, KE Leslie, JS Walton, SJ LeBlanc (2010) Risk factors for postpartum uterine diseases in dairy cows. J Dairy Sci 93: 5764-5771.
- Williams EJ, DP Fischer, DU Pfeiffer, GCW England, DE Noakes, et al. (2005) Clinical evaluation of postpartum vaginal mucus reflects uterine bacterial infection and the immune response in cattle. Theriogenology 63: 102-117.
- Wagener K, T Grunert, I Prunner, M Ehling-Schulz, M Drillich (2014) Dynamics of uterine infections with Escherichia coli, Streptococcus uberis and Trueperella pyogenes in post-partum dairy cows and their association with clinical endometritis. Vet J 202: 527-532.
- Bicalho MLS, FS Lima, VS Machado, EB Meira Jr, EK Ganda, et al. (2016) Associations among Trueperella pyogenes, endometritis diagnosis, and pregnancy outcomes in dairy cows. Theriogenology 85: 267-274.
- Bicalho RC, VS Machado, MLS Bicalho, RO Gilbert, AGV Teixeira, et al. (2010) Molecular and epidemiological characterization of bovine intrauterine Escherichia coli. J Dairy Sci 93: 5818-5830.
- Prunner I, H Pothmann, K Wagener, M Giuliodori, J Huber, et al. (2014) Dynamics of bacteriologic and cytologic changes in the uterus of postpartum dairy cows. Theriogenology 82: 1316-1322.
- Kaper JB, JP Nataro, HL Mobley (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2: 123-140.
- Sheldon IM, AN Rycroft, B Dogan, M Craven, JJ Bromfield, et al. (2010) Specific strains of Escherichia coli are pathogenic for the endometrium of cattle and cause pelvic inflammatory disease in cattle and mice. PLoS One 5: e9192.
- Silva E, S Leitão, T Tenreiro, C Pomba, T Nunes, et al. (2009) Genomic and phenotypic characterization of Escherichia coli isolates recovered from the uterus of puerperal dairy cows. J Dairy Sci 92: 6000-6010.
- Kassé FN, JM Fairbrother, J Dubuc (2016) Relationship between Escherichia coli virulence factors and postpartum metritis in dairy cows. J Dairy Sci 99: 4656-4667.
- Dohoo IR, SW Martin, H Stryhn (2009) Veterinary Epidemiologic Research. (2nd edn). VER Inc., Charlottetown, PEI, Canada.
- Maluta RP, JM Fairbrother, AE Stella, EC Rigobelo, R Marinez, et al. (2014) Potentially pathogenic Escherichia coli in healthy, pasture-raised sheep on farms and at the abattoir in Brazil. Vet Microbiol 169: 89-95.
- Fairbrother JH, S Dufour, JM Fairbrother, D Francoz, E Nadeau, et al. (2015) Characterization of persistent and transient Escherichia coli isolates recovered from clinical mastitis episodes in dairy cows. Vet Microbiol 176: 126-133.
- Kasimanickam R, TF Duffield, RA Foster, CJ Gartley, KE Leslie, et al. (2004) Endometrial cytology and ultrasonography for the detection of subclinical endometritis in postpartum dairy cows 62: 9-23.
- Litwin CM, SB Calderwood (1993) Role of Iron in Regulation of Virulence Genes. Clin Microbiol Rev 6: 137-149.
- Autenrieth I, K Hantke, J Heesemann (1991) Immunosuppression of the host and delivery of iron to the pathogen: a possible dual role of siderophores in the pathogenesis of microbial infections? Med Microbiol Immunol 180: 135-141.
- Schubert S, A Rakin, H Karch, E Carniel, J Heesemann (1998) Prevalence of the “High-Pathogenicity Island” of Yersinia Species among Escherichia coli Strains That Are Pathogenic to Humans. Infect Immun 66(2): 480- 485.
- Dezfulian H, I Batisson, JM Fairbrother, PCK Lau, A Nassar, et al. (2003) Presence and Characterization of Extraintestinal Pathogenic Escherichia coli Virulence Genes in F165-Positive E. coli Strains Isolated from Diseased Calves and Pigs. J Clin Microbiol 41: 1375-1385
- Tourret J, M Diard, L Garry, I Matic, E Denamur (2010) Effects of single and multiple pathogenicity island deletions on uropathogenic Escherichia coli strain 536 intrinsic extra-intestinal virulence. Internat J Med Microbiol 300: 435-439.
- Johnson JR, E Oswald, TT O’Bryan, MA Kuskowski, L Spanjaard (2002) Phylogenetic Distribution of Virulence-Associated Genes among Escherichia coli Isolates Associated with Neonatal Bacterial Meningitis in The Netherlands. J Infect Diseases 185: 774-784.
- Johnson JR, MA Kuskowski, A Gajewski, S Soto, JP Horcajada, et al. (2005) Extended Virulence Genotypes and Phylogenetic Background of Escherichia coli Isolates from Patients with Cystitis, Pyelonephritis, or Prostatitis. J Infect Diseases 191: 46-50.
- Östblom A, I Adlerberth, AE Wold, FL Nowrouzian (2011) Pathogenicity Island Markers, Virulence Determinants malX and usp, and the Capacity of Escherichia coli To Persist in Infants’ Commensal Microbiotas. Appl Environm Microbiol 77: 2303-2308.
- Lynne AM, JA Skyberg, CM Logue, C Doetkott, SL Foley, et al. (2007) Characterization of a Series of Transconjugant Mutants of an Avian Pathogenic Escherichia coli Isolate for Resistance to Serum Complement. Avian Diseases 51: 771-776.
- Chuba PJ, MA Leon, A Banerjee, S Palehaudhuri (1989) Cloning and DNA sequence of plasmid determinant iss, coding for increased serum survival and surface exclusion, which has homology with lambda DNA. Mol Gen Genet 216: 287-292.
- Johnson JR (1991) Virulence Factors in Escherichia coli Urinary Tract Infection. Clin Microbiol Rev 4: 80-128.
- Griffin JFT, PJ Hartigan, WR Nunn (1974) Non-specific uterine infection and bovine fertility: I. Infection patterns and endometritis during the first seven weeks post-partum. Theriogenology 1: 91-106.
- Sheldon IM, DE Noakes, AN Rycroft, DU Pfeiffer, H Dobson (2002) Influence of uterine bacterial contamination after parturition on ovarian dominant follicle selection and follicle growth and function in cattle. Reprod 123: 837-845.
- Santos TMA, RO Gilbert, RC Bicalho (2011) Metagenomic analysis of the uterine bacterial microbiota in healthy and metritic postpartum dairy cows. 94: 291-302.
- Sheldon IM, J Cronin, L Goetze, G Donofrio, HJ Schuberth (2009) Defining Postpartum Uterine Disease and the Mechanisms of Infection and Immunity in the Female Reproductive Tract in Cattle. Biol Reprod 81: 1025-1032.
- An H, J M Fairbrother, C Desautels, J Harel (1999) Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv Exp Med Biol 473: 179-184.
- Bauer RJ, L Zhang, B Foxman, A Siitonen, ME Jantunen, et al. (2002) Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection-usp, iha, and iroN (E. coli). J Infect Dis 185: 1521-1524.
- Beaudry M, C Zhu, JM Fairbrother, J Harel (1996) Genotypic and phenotypic characterization of Escherichia coli isolates from dogs manifesting attaching and effacing lesions. J Clin Microbiol 34: 144-148.
- Chapman TA, XY Wu, I Barchia, KA Bettelheim, S Driesen, et al. (2006) Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl Environ Microbiol 72: 4782- 4795.
- Cid D, R Sanz, I Marin, H de Greve, JA Ruiz-Santa-Quiteria, et al. (1999) Characterization of non-enterotoxigenic Escherichia coli strains producing F17 fimbriae isolated from diarrheic lambs and goat kids. J Clin Microbiol 37: 1370-1375.
- Daigle F, J Harel, JM Fairbrother, P Lebel (1994) Expression and detection of pap-, sfa-, and afa-encoded fimbrial adhesin systems among uropathogenic Escherichia coli. Can. J. Microbiol. 40: 286-291.
- Dozois CM, M Dho Moulin, A Bree, JM Fairbrother, C Desautels, et al. (2000) Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect Immun 68:4145-4154.
- Ewers C, G Li, H Wilking, S Kiessling, K Alt, et al. (2007) Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int J Med Microbiol 297: 163-176.
- Furrer B, U Candrian, J Luthy (1990) Detection and identification of E. coli producing heat-labile enterotoxin type I by enzymatic amplification of a specific DNA fragment. Lett Appl Microbiol 10: 31-34.
- Girardeau JP, Y Bertin, C Martin, M Der Vartanian, C Boeuf (1991) Sequence analysis of the clpG gene, which codes for surface antigen CS31A subunit: Evidence of an evolutionary relationship between CS31A, K88, and F41 subunit genes. J Bacteriol 173: 7673-7683.
- Goluszko P, SL Moseley, LD Truong, A Kaul, JR Williford, et al. (1997) Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae: Mutation in the dra region prevented tubulointerstitial nephritis. J Clin Invest 99: 1662- 1672.
- Herrero M, V de Lorenzo, JB Neilands (1988) Nucleotide sequence of the iucD gene of the pColV-K30 aerobactin operon and topology of its product studied with phoA and lacZ gene fusions. J Bacteriol 170: 56-64.
- Hilali F, R Ruimy, P Saulnier, C Barnabe, C Lebouguenec, et al. (2000) Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect Immun 68: 3983-3989.
- Karch H, S Schubert, D Zhang, W Zhang, H Schmidt, et al. (1999) A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun 67: 5994-6001.
- Lortie LA, JD Dubreuil, J Harel (1991) Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. J Clin Microbiol 29: 656- 659. Maldonado G, S Greenland (1993) Simulation study of confounder-selection strategies. Am J Epidemiol 138: 923-936.
- McVey DS, M Kennedy, MM Chengappa (2013) Veterinary Microbiology. (3rd edn) Chapter 28, page 203. John Wiley and Sons Inc., Ames, IA.
- Ngeleka M, J Pritchard, G Appleyard, DM Middleton, JM Fairbrother (2003) Isolation and association of Escherichia coli AIDA-I/STb, rather than EAST1 pathotype, with diarrhea in piglets and antibiotic sensitivity of isolates. J Vet Diagn Invest 15: 242-252.
- Ojeniyi B, P Ahrens, A Meyling (1994) Detection of fimbrial and toxin genes in Escherichia coli and their prevalence in piglets with diarrhoea. The application of colony hybridization assay, polymerase chain reaction and phenotypic assays. Zentralbl Veterinarmed B 41: 49-59.
- Oswald E, M Tabouret, R Boivin, J De Rycke (1994) Detection of Escherichia coli strains producing cytotoxic necrotizing factor type two (CNF2) by enzyme-linked immunosorbent assay. Vet Microbiol 40: 209- 218.
- Rodriguez Siek KE, CW Giddings, C Doetkott, TJ Johnson, LK Nolan (2005) Characterizing the APEC pathotype. Vet Res 36: 241-256.
- Sandhu KS, RC Clarke, CL Gyles (1997) Hemolysin phenotypes and genotypes of eaeA-positive and eaeA-negative bovine verotoxigenic Escherichia coli. Adv Exp Med Biol 412: 295-302.
- Savarino SJ, A Fasano, J Watson, BM Martin, MM Levine, et al. (1993) Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci USA 90: 3093-3097.
- So M, BJ McCarthy (1980) Nucleotide sequence of the bacterial transposon Tn1681 encoding a heat-stable (ST) toxin and its identification in enterotoxigenic Escherichia coli strains. Proc Natl Acad Sci USA 77: 4011-4015.
- Woodward MJ, PJ Carroll, C Wray (1992) Detection of entero- and verocyto-toxin genes in Escherichia coli from diarrhoeal disease in animals using the polymerase chain reaction. Vet Microbiol 31: 251-261.
- Ye C, J Xu (2001) Prevalence of iron transport gene on pathogenicityassociated island of uropathogenic Escherichia coli in E. coli O157:H7 containing Shiga toxin gene. J Clin Microbiol 39: 2300-2305.

Top Editors
-

Mark E Smith
Bio chemistry
University of Texas Medical Branch, USA -

Lawrence A Presley
Department of Criminal Justice
Liberty University, USA -

Thomas W Miller
Department of Psychiatry
University of Kentucky, USA -

Gjumrakch Aliev
Department of Medicine
Gally International Biomedical Research & Consulting LLC, USA -

Christopher Bryant
Department of Urbanisation and Agricultural
Montreal university, USA -

Robert William Frare
Oral & Maxillofacial Pathology
New York University, USA -

Rudolph Modesto Navari
Gastroenterology and Hepatology
University of Alabama, UK -

Andrew Hague
Department of Medicine
Universities of Bradford, UK -

George Gregory Buttigieg
Maltese College of Obstetrics and Gynaecology, Europe -

Chen-Hsiung Yeh
Oncology
Circulogene Theranostics, England -
.png)
Emilio Bucio-Carrillo
Radiation Chemistry
National University of Mexico, USA -
.jpg)
Casey J Grenier
Analytical Chemistry
Wentworth Institute of Technology, USA -
Hany Atalah
Minimally Invasive Surgery
Mercer University school of Medicine, USA -

Abu-Hussein Muhamad
Pediatric Dentistry
University of Athens , Greece

The annual scholar awards from Lupine Publishers honor a selected number Read More...




